[English] 日本語
 Yorodumi
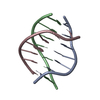
Yorodumi- PDB-177d: SOLUTION STRUCTURE AND HYDRATION PATTERNS OF A PYRIMIDINE(DOT)PUR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 177d | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE AND HYDRATION PATTERNS OF A PYRIMIDINE(DOT)PURINE(DOT)PYRIMIDINE DNA TRIPLEX CONTAINING A NOVEL T(DOT)CG TRIPLE | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / TRIPLEX | Function / homology | DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / MOLECULAR DYNAMICS, DISTANCE GEOMETRY |  Authors AuthorsRadhakrishnan, I. / Patel, D.J. |  Citation Citation Journal: J.Mol.Biol. / Year: 1994 Journal: J.Mol.Biol. / Year: 1994Title: Solution structure and hydration patterns of a pyrimidine.purine.pyrimidine DNA triplex containing a novel T.CG base-triple. Authors: Radhakrishnan, I. / Patel, D.J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  177d.cif.gz 177d.cif.gz | 100.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb177d.ent.gz pdb177d.ent.gz | 67.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  177d.json.gz 177d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/77/177d https://data.pdbj.org/pub/pdb/validation_reports/77/177d ftp://data.pdbj.org/pub/pdb/validation_reports/77/177d ftp://data.pdbj.org/pub/pdb/validation_reports/77/177d | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 9407.036 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: CHEMICALLY SYNTHESIZED /  Keywords: DEOXYRIBONUCLEIC ACID Keywords: DEOXYRIBONUCLEIC ACID |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Developer: BRUNGER / Classification: refinement X-PLOR / Developer: BRUNGER / Classification: refinement | ||||||||
| Refinement | Method: MOLECULAR DYNAMICS, DISTANCE GEOMETRY / Software ordinal: 1 Details: NUMBER OF NUCLEIC ACID ATOMS -- 415 RESTRAINED MOLECULAR DYNAMICS CALCULATIONS WERE CONDUCTED ON IDEALIZED A'- AND B-FORM STARTING STRUCTURES. ONLY THE TRIPLEX REGION IN THE SEQUENCE WAS ...Details: NUMBER OF NUCLEIC ACID ATOMS -- 415 RESTRAINED MOLECULAR DYNAMICS CALCULATIONS WERE CONDUCTED ON IDEALIZED A'- AND B-FORM STARTING STRUCTURES. ONLY THE TRIPLEX REGION IN THE SEQUENCE WAS CONSIDERED. SIX STRUCTURES WERE CALCULATED INITIALLY USING DISTANCE RESTRAINTS. A MAJORITY OF THE DISTANCE RESTRAINTS WERE SUBSEQUENTLY REPLACED BY NOE INTENSITY RESTRAINTS. THE R(1/6) VALUE WAS MONITORED DURING THE REFINEMENTS. EACH STRUCTURE WAS SUBJECTED TO 500 CYCLES OF ENERGY MINIMIZATION AFTER THE DYNAMICS. NO AVERAGE STRUCTURE WAS CALCULATED. THE R(1/6) VALUE FOR THE MINIMIZED STRUCTURE REPRESENTING THE ENSEMBLE IS 0.024. THE DYNAMICS SIMULATIONS WERE PERFORMED IN THE PRESENCE OF EXPLICIT SOLVENT MOLECULES AND 15 NA+ COUNTER IONS. THE SUMMATION EXTENDS THROUGH ALL ASSIGNABLE, QUANTIFIABLE CROSS PEAK INTENSITIES IN NOESY SPECTRA RECORDED AT MIXING TIMES OF 40, 90 AND 150 MS. RMS DEVIATIONS FROM IDEALIZED GEOMETRY FOR THE MINIMIZED STRUCTURE `RM' REPRESENTING THE ENSEMBLE ARE AS FOLLOWS: RM BOND (ANGSTROMS) 0.016 (0.016) ANGLES (DEGREES) 3.825 (3.734) IMPROPERS (DEGREES) 0.552 (0.391) THE VALUES IN PARENTHESES CORRESPOND TO THE AVERAGE VALUES FOR THE ENSEMBLE OF SIX STRUCTURES. | ||||||||
| NMR ensemble | Conformer selection criteria: all calculated structures submitted Conformers calculated total number: 6 / Conformers submitted total number: 6 |
 Movie
Movie Controller
Controller







 PDBj
PDBj






































