[English] 日本語
 Yorodumi
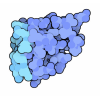
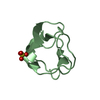
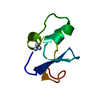
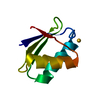
Yorodumi- PDB-1ops: ICE-BINDING SURFACE ON A TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ops | ||||||
|---|---|---|---|---|---|---|---|
| Title | ICE-BINDING SURFACE ON A TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT | ||||||
 Components Components | TYPE III ANTIFREEZE PROTEIN | ||||||
 Keywords Keywords | ANTIFREEZE PROTEIN / ICE CRYSTAL GROWTH INHIBITION / PRETZEL FOLD / GLYCOPROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Macrozoarces americanus (ocean pout) Macrozoarces americanus (ocean pout) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Yang, D.S.C. / Hon, W.-C. / Bubanko, S. / Xue, Y. / Seetharaman, J. / Hew, C.L. / Sicheri, F. | ||||||
 Citation Citation |  Journal: Biophys.J. / Year: 1998 Journal: Biophys.J. / Year: 1998Title: Identification of the ice-binding surface on a type III antifreeze protein with a "flatness function" algorithm. Authors: Yang, D.S. / Hon, W.C. / Bubanko, S. / Xue, Y. / Seetharaman, J. / Hew, C.L. / Sicheri, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ops.cif.gz 1ops.cif.gz | 22.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ops.ent.gz pdb1ops.ent.gz | 13.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ops.json.gz 1ops.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/op/1ops https://data.pdbj.org/pub/pdb/validation_reports/op/1ops ftp://data.pdbj.org/pub/pdb/validation_reports/op/1ops ftp://data.pdbj.org/pub/pdb/validation_reports/op/1ops | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6750.958 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: ISOFORM HPLC-3 / Source: (natural)  Macrozoarces americanus (ocean pout) / Tissue: SERUM / References: UniProt: P19608 Macrozoarces americanus (ocean pout) / Tissue: SERUM / References: UniProt: P19608 |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.05 Å3/Da / Density % sol: 40.05 % Description: THE STRUCTURE WAS SOLVED BY THE METHOD OF ITERATIVE SINGLE ANOMALOUS SCATTERING (B.C. WANG, (1985), METHODS ENZYMOL., 114, P90-117). | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 8 / Details: pH 8.0 | ||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 277 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: RIGAKU / Detector: IMAGE PLATE / Date: May 1, 1993 / Details: SUPPER DOUBLE FOCUSING MIRRORS |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2→40 Å / Num. obs: 3107 / % possible obs: 82.4 % / Observed criterion σ(I): 2 / Biso Wilson estimate: 19.3 Å2 / Rsym value: 0.066 |
| Reflection | *PLUS Rmerge(I) obs: 0.066 |
| Reflection shell | *PLUS % possible obs: 54.9 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: THE STARTING STRUCTURAL MODEL WAS DETERMINED FROM A CRYSTAL GROWN WITH THE RECOMBINANT, SINGLY-IODINATED PROTEIN, ISOFORM HPLC-6. THIS MODEL IS NOT YET AVAILABLE. Resolution: 2→40 Å / Rfactor Rfree error: 0.016 / Data cutoff high absF: 10000000 / Data cutoff low absF: 0.001 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 2 / Details: BULK ENT MODEL USED
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.4 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→40 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.07 Å / Rfactor Rfree error: 0.06 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.843 / Classification: refinement X-PLOR / Version: 3.843 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2 Å / Lowest resolution: 40 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.318 |
 Movie
Movie Controller
Controller









 PDBj
PDBj