+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9906 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of membrane protein | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | PTS / ManYZ / transporter / Mannose / PROTEIN TRANSPORT | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmannose transmembrane transport / protein-N(PI)-phosphohistidine-mannose phosphotransferase system transporter activity / fructose import across plasma membrane / N-acetylglucosamine transport / D-glucose import across plasma membrane / phosphoenolpyruvate-dependent sugar phosphotransferase system / transmembrane transporter complex / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |    | ||||||||||||
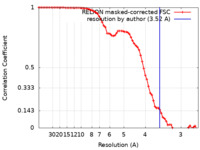
| Method | single particle reconstruction / cryo EM / Resolution: 3.52 Å | ||||||||||||
 Authors Authors | Wang JW / Zeng JW | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2019 Journal: Cell Res / Year: 2019Title: Structure of the mannose transporter of the bacterial phosphotransferase system. Authors: Xueli Liu / Jianwei Zeng / Kai Huang / Jiawei Wang /  | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9906.map.gz emd_9906.map.gz | 19.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9906-v30.xml emd-9906-v30.xml emd-9906.xml emd-9906.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9906_fsc.xml emd_9906_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_9906.png emd_9906.png | 63.7 KB | ||
| Filedesc metadata |  emd-9906.cif.gz emd-9906.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9906 http://ftp.pdbj.org/pub/emdb/structures/EMD-9906 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9906 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9906 | HTTPS FTP |
-Related structure data
| Related structure data |  6k1hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9906.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9906.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.338 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ManYZ trimer complex
| Entire | Name: ManYZ trimer complex |
|---|---|
| Components |
|
-Supramolecule #1: ManYZ trimer complex
| Supramolecule | Name: ManYZ trimer complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 176 kDa/nm |
-Macromolecule #1: PTS mannose transporter subunit IID
| Macromolecule | Name: PTS mannose transporter subunit IID / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.983373 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVDTTQTTTE KKLTQSDIRG VFLRSNLFQG SWNFERMQAL GFCFSMVPAI RRLYPENNEA RKQAIRRHLE FFNTQPFVAA PILGVTLAL EEQRANGAEI DDGAINGIKV GLMGPLAGVG DPIFWGTVRP VFAALGAGIA MSGSLLGPLL FFILFNLVRL A TRYYGVAY ...String: MVDTTQTTTE KKLTQSDIRG VFLRSNLFQG SWNFERMQAL GFCFSMVPAI RRLYPENNEA RKQAIRRHLE FFNTQPFVAA PILGVTLAL EEQRANGAEI DDGAINGIKV GLMGPLAGVG DPIFWGTVRP VFAALGAGIA MSGSLLGPLL FFILFNLVRL A TRYYGVAY GYSKGIDIVK DMGGGFLQKL TEGASILGLF VMGALVNKWT HVNIPLVVSR ITDQTGKEHV TTVQTILDQL MP GLVPLLL TFACMWLLRK KVNPLWIIVG FFVIGIAGYA CGLLGL UniProtKB: UNIPROTKB: A0A387CW08 |
-Macromolecule #2: PTS system mannose-specific EIIC component
| Macromolecule | Name: PTS system mannose-specific EIIC component / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.752783 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEITTLQIVL VFIVACIAGM GSILDEFQFH RPLIACTLVG IVLGDMKTGI IIGGTLEMIA LGWMNIGAAV APDAALASII STILVIAGH QSIGAGIALA IPLAAAGQVL TIIVRTITVA FQHAADKAAD NGNLTAISWI HVSSLFLQAM RVAIPAVIVA L SVGTSEVQ ...String: MEITTLQIVL VFIVACIAGM GSILDEFQFH RPLIACTLVG IVLGDMKTGI IIGGTLEMIA LGWMNIGAAV APDAALASII STILVIAGH QSIGAGIALA IPLAAAGQVL TIIVRTITVA FQHAADKAAD NGNLTAISWI HVSSLFLQAM RVAIPAVIVA L SVGTSEVQ NMLNAIPEVV TNGLNIAGGM IVVVGYAMVI NMMRAGYLMP FFYLGFVTAA FTNFNLVALG VIGTVMAVLY IQ LSPKYNR VAGAPAQAAG NNDLDNELDH HHHHHHH UniProtKB: PTS system mannose-specific EIIC component |
-Macromolecule #3: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 3 / Number of copies: 3 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)