[English] 日本語
 Yorodumi
Yorodumi- EMDB-5263: Quaternary structures of HIV Env immunogen exhibit conformational... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5263 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding | |||||||||
 Map data Map data | gp140 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / Env trimer / cryoelectron microscopy / induced conformation | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 19.0 Å | |||||||||
 Authors Authors | Moscoso CG / Sun Y / Poon S / Xing L / Kan E / Martin L / Green DJ / Lin F / Vahlne A / Barnett SW ...Moscoso CG / Sun Y / Poon S / Xing L / Kan E / Martin L / Green DJ / Lin F / Vahlne A / Barnett SW / Srivastava IK / Cheng RH | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2011 Journal: Proc Natl Acad Sci U S A / Year: 2011Title: Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Authors: Carlos G Moscoso / Yide Sun / Selina Poon / Li Xing / Elaine Kan / Loïc Martin / Dominik Green / Frank Lin / Anders G Vahlne / Susan Barnett / Indresh Srivastava / R Holland Cheng /  Abstract: The human immunodeficiency virus envelope protein is the key element mediating entry into host cells. Conformational rearrangement of Env upon binding to the host CD4 receptor and chemokine ...The human immunodeficiency virus envelope protein is the key element mediating entry into host cells. Conformational rearrangement of Env upon binding to the host CD4 receptor and chemokine coreceptor drives membrane fusion. We elucidated the quaternary arrangement of the soluble Env trimeric immunogen o-gp140ΔV2TV1, in both its native (unliganded) and CD4-induced (liganded) states by cryoelectron microscopy and molecular modeling. The liganded conformation was elicited by binding gp140 to the synthetic CD4-mimicking miniprotein CD4m. Upon CD4m binding, an outward domain shift of the three gp120 subunits diminishes gp120-gp41 interactions, whereas a "flat open" concave trimer apex is observed consequent to gp120 tilting away from threefold axis, likely juxtaposing the fusion peptide with the host membrane. Additional features observed in the liganded conformation include rotations of individual gp120 subunits that may release gp41 for N- and C-helix refolding and also may lead to optimal exposure of the elicited coreceptor binding site. Such quaternary arrangements of gp140 lead to the metastable liganded conformation, with putative locations of exposed epitopes contributing to a description of sequential events occurring prior to membrane fusion. Our observations imply a mechanism whereby a soluble Env trimeric construct, as opposed to trimers extracted from virions, may better expose crucial epitopes such as the CD4 binding site and V3, as well as epitopes in the vicinity of gp41, subsequent to conjugation with CD4m. Structural features gleaned from our studies should aid the design of Env-based immunogens for inducement of potent broadly neutralizing antibodies against exposed conformational epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5263.map.gz emd_5263.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5263-v30.xml emd-5263-v30.xml emd-5263.xml emd-5263.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5263_1.tif emd_5263_1.tif | 109.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5263 http://ftp.pdbj.org/pub/emdb/structures/EMD-5263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5263 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5263.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5263.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | gp140 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
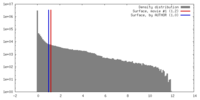
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : o-gp140dV2TV1 trimer
| Entire | Name: o-gp140dV2TV1 trimer |
|---|---|
| Components |
|
-Supramolecule #1000: o-gp140dV2TV1 trimer
| Supramolecule | Name: o-gp140dV2TV1 trimer / type: sample / ID: 1000 / Oligomeric state: Trimer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 450 KDa / Theoretical: 450 KDa / Method: Triple detector array system |
-Macromolecule #1: o-gp140dV2TV1
| Macromolecule | Name: o-gp140dV2TV1 / type: protein_or_peptide / ID: 1 / Name.synonym: gp140 / Number of copies: 3 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / synonym: HIV-1 Human immunodeficiency virus 1 / synonym: HIV-1 |
| Molecular weight | Experimental: 450 KDa / Theoretical: 450 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.9 / Details: 20 mM Tris, 50 mM NaCl |
| Grid | Details: 200 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 93 K / Instrument: REICHERT-JUNG PLUNGER / Details: Vitrification instrument: Reichert plunger / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Temperature | Average: 100 K |
| Date | Jul 23, 2008 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Digitization - Sampling interval: 15 µm / Number real images: 101 / Average electron dose: 15 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 80000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: SITUS |
| Details | PDBEntryID_givenInChain. Protocol: Rigid Body. The coordinates were initially manually fitted using program Chimera and refined using program SITUS |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)