登録情報 データベース : EMDB / ID : EMD-9905タイトル RSC substrate-recruitment module RSC SRM region / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Saccharomyces cerevisiae S288c (パン酵母) / Saccharomyces cerevisiae S288C (パン酵母)手法 / / 解像度 : 3.4 Å Ye YP / Wu H ジャーナル : Science / 年 : 2019タイトル : Structure of the RSC complex bound to the nucleosome.著者 : Youpi Ye / Hao Wu / Kangjing Chen / Cedric R Clapier / Naveen Verma / Wenhao Zhang / Haiteng Deng / Bradley R Cairns / Ning Gao / Zhucheng Chen / 要旨 : The RSC complex remodels chromatin structure and regulates gene transcription. We used cryo-electron microscopy to determine the structure of yeast RSC bound to the nucleosome. RSC is delineated into ... The RSC complex remodels chromatin structure and regulates gene transcription. We used cryo-electron microscopy to determine the structure of yeast RSC bound to the nucleosome. RSC is delineated into the adenosine triphosphatase motor, the actin-related protein module, and the substrate recruitment module (SRM). RSC binds the nucleosome mainly through the motor, with the auxiliary subunit Sfh1 engaging the H2A-H2B acidic patch to enable nucleosome ejection. SRM is organized into three substrate-binding lobes poised to bind their respective nucleosomal epitopes. The relative orientations of the SRM and the motor on the nucleosome explain the directionality of DNA translocation and promoter nucleosome repositioning by RSC. Our findings shed light on RSC assembly and functionality, and they provide a framework to understand the mammalian homologs BAF/PBAF and the Sfh1 ortholog INI1/BAF47, which are frequently mutated in cancers. 履歴 登録 2019年5月9日 - ヘッダ(付随情報) 公開 2019年11月13日 - マップ公開 2019年11月13日 - 更新 2024年3月27日 - 現状 2024年3月27日 処理サイト : PDBj / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
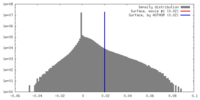
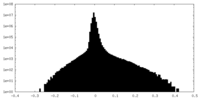
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報

 データ登録者
データ登録者 引用
引用 ジャーナル: Science / 年: 2019
ジャーナル: Science / 年: 2019

 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_9905.map.gz
emd_9905.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-9905-v30.xml
emd-9905-v30.xml emd-9905.xml
emd-9905.xml EMDBヘッダ
EMDBヘッダ emd_9905.png
emd_9905.png emd-9905.cif.gz
emd-9905.cif.gz emd_9905_additional_1.map.gz
emd_9905_additional_1.map.gz emd_9905_additional_2.map.gz
emd_9905_additional_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-9905
http://ftp.pdbj.org/pub/emdb/structures/EMD-9905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9905
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9905 emd_9905_validation.pdf.gz
emd_9905_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_9905_full_validation.pdf.gz
emd_9905_full_validation.pdf.gz emd_9905_validation.xml.gz
emd_9905_validation.xml.gz emd_9905_validation.cif.gz
emd_9905_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9905
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9905 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9905
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9905 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_9905.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_9905.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析 ムービー
ムービー コントローラー
コントローラー






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)