+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9632 | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of Enterovirus D68 procapsid | ||||||||||||||||||||||||||||||||||||
 Map data Map data | None | ||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Enterovirus D68 / procapsid / VIRUS | ||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell membrane / cysteine-type peptidase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / helicase activity / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity ...host cell membrane / cysteine-type peptidase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / helicase activity / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / viral capsid / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / host cell cytoplasm / RNA helicase activity / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||||||||||||||||||||||||||||||||
| Biological species |  Enterovirus D68 / Enterovirus D68 /  Enterovirus d Enterovirus d | ||||||||||||||||||||||||||||||||||||
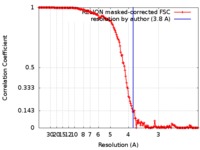
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Zheng QB / Zhu R | ||||||||||||||||||||||||||||||||||||
| Funding support |  China, China,  United States, 11 items United States, 11 items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2019 Journal: Nat Microbiol / Year: 2019Title: Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization. Authors: Qingbing Zheng / Rui Zhu / Longfa Xu / Maozhou He / Xiaodong Yan / Dongxiao Liu / Zhichao Yin / Yangtao Wu / Yongchao Li / Lisheng Yang / Wangheng Hou / Shuxuan Li / Zizhen Li / Zhenqin Chen ...Authors: Qingbing Zheng / Rui Zhu / Longfa Xu / Maozhou He / Xiaodong Yan / Dongxiao Liu / Zhichao Yin / Yangtao Wu / Yongchao Li / Lisheng Yang / Wangheng Hou / Shuxuan Li / Zizhen Li / Zhenqin Chen / Zhihai Li / Hai Yu / Ying Gu / Jun Zhang / Timothy S Baker / Z Hong Zhou / Barney S Graham / Tong Cheng / Shaowei Li / Ningshao Xia /   Abstract: Enterovirus D68 (EV-D68) undergoes structural transformation between mature, cell-entry intermediate (A-particle) and empty forms throughout its life cycle. Structural information for the various ...Enterovirus D68 (EV-D68) undergoes structural transformation between mature, cell-entry intermediate (A-particle) and empty forms throughout its life cycle. Structural information for the various forms and antibody-bound capsids will facilitate the development of effective vaccines and therapeutics against EV-D68 infection, which causes childhood respiratory and paralytic diseases worldwide. Here, we report the structures of three EV-D68 capsid states representing the virus at major phases. We further describe two original monoclonal antibodies (15C5 and 11G1) with distinct structurally defined mechanisms for virus neutralization. 15C5 and 11G1 engage the capsid loci at icosahedral three-fold and five-fold axes, respectively. To block viral attachment, 15C5 binds three forms of capsids, and triggers mature virions to transform into A-particles, mimicking engagement by the functional receptor ICAM-5, whereas 11G1 exclusively recognizes the A-particle. Our data provide a structural and molecular explanation for the transition of picornavirus capsid conformations and demonstrate distinct mechanisms for antibody-mediated neutralization. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9632.map.gz emd_9632.map.gz | 324.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9632-v30.xml emd-9632-v30.xml emd-9632.xml emd-9632.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9632_fsc.xml emd_9632_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_9632.png emd_9632.png | 290.6 KB | ||
| Filedesc metadata |  emd-9632.cif.gz emd-9632.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9632 http://ftp.pdbj.org/pub/emdb/structures/EMD-9632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9632 | HTTPS FTP |
-Related structure data
| Related structure data |  6aj3MC  9629C  9631C  9633C  9634C  9635C  9636C  6aj0C  6aj2C  6aj7C  6aj9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9632.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9632.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.128 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Enterovirus d
| Entire | Name:  Enterovirus d Enterovirus d |
|---|---|
| Components |
|
-Supramolecule #1: Enterovirus d
| Supramolecule | Name: Enterovirus d / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 42769 / Sci species name: Enterovirus d / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 Enterovirus D68 |
| Molecular weight | Theoretical: 32.673035 KDa |
| Sequence | String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSNTEPE EAIQTRTVIN QHGVSETLVE NFLSRAALVS KRSFEYKDHT SSAAQADKN FFKWTINTRS FVQLRRKLEL FTYLRFDAEI TILTTVAVNG SGNNTYVGLP DLTLQAMFVP TGALTPEKQD S FHWQSGSN ...String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSNTEPE EAIQTRTVIN QHGVSETLVE NFLSRAALVS KRSFEYKDHT SSAAQADKN FFKWTINTRS FVQLRRKLEL FTYLRFDAEI TILTTVAVNG SGNNTYVGLP DLTLQAMFVP TGALTPEKQD S FHWQSGSN ASVFFKISDP PARITIPFMC INSAYSVFYD GFAGFEKNGL YGINPADTIG NLCVRIVNEH QPVGFTVTVR VY MKPKHIK AWAPRPPRTL PYMSIANANY KGKERAPNAL NAIIGNRDSV KTMPHNIV UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 Enterovirus D68 |
| Molecular weight | Theoretical: 27.567135 KDa |
| Sequence | String: SPSAEACGYS DRVLQLKLGN SAIVTQEAAN YCCAYGEWPN YLPDHEAVAI DKPTQPETAT DRFYTLKSVK WETGSTGWWW KLPDALNNI GMFGQNVQHH YLYRSGFLIH VQCNATKFHQ GALLVVAIPE HQRGAHNTNT SPGFDDIMKG EEGGTFNHPY V LDDGTSLA ...String: SPSAEACGYS DRVLQLKLGN SAIVTQEAAN YCCAYGEWPN YLPDHEAVAI DKPTQPETAT DRFYTLKSVK WETGSTGWWW KLPDALNNI GMFGQNVQHH YLYRSGFLIH VQCNATKFHQ GALLVVAIPE HQRGAHNTNT SPGFDDIMKG EEGGTFNHPY V LDDGTSLA CATIFPHQWI NLRTNNSATI VLPWMNAAPM DFPLRHNQWT LAIIPVVPLG TRTTSSMVPI TVSIAPMCCE FN GLRHAIT Q UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 Enterovirus D68 |
| Molecular weight | Theoretical: 27.142842 KDa |
| Sequence | String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNMLEVVQVE SMMEINNTES AVGMERLKVD ISALTDVDQL LFNIPLDIQ LDGPLRNTLV GNISRYYTHW SGSLEMTFMF CGSFMATGKL ILCYTPPGGS CPTTRETAML GTHIVWDFGL Q SSVTLIIP ...String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNMLEVVQVE SMMEINNTES AVGMERLKVD ISALTDVDQL LFNIPLDIQ LDGPLRNTLV GNISRYYTHW SGSLEMTFMF CGSFMATGKL ILCYTPPGGS CPTTRETAML GTHIVWDFGL Q SSVTLIIP WISGSHYRMF NNDAKSTNAN VGYVTCFMQT NLIVPSESSD TCSLIGFIAA KDDFSLRLMR DSPDIGQLDH LH AAEAAYQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)