+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9346 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli ATP synthase after incubation with ATP - State B | |||||||||
 Map data Map data | E. coli ATP synthase after incubation with ATP | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.4 Å | |||||||||
 Authors Authors | Sobit M / Stewart AG | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Cryo-EM reveals distinct conformations of ATP synthase on exposure to ATP. Authors: Meghna Sobti / Robert Ishmukhametov / James C Bouwer / Anita Ayer / Cacang Suarna / Nicola J Smith / Mary Christie / Roland Stocker / Thomas M Duncan / Alastair G Stewart /    Abstract: ATP synthase produces the majority of cellular energy in most cells. We have previously reported cryo-EM maps of autoinhibited ATP synthase imaged without addition of nucleotide (Sobti et al. 2016), ...ATP synthase produces the majority of cellular energy in most cells. We have previously reported cryo-EM maps of autoinhibited ATP synthase imaged without addition of nucleotide (Sobti et al. 2016), indicating that the subunit ε engages the α, β and γ subunits to lock the enzyme and prevent functional rotation. Here we present multiple cryo-EM reconstructions of the enzyme frozen after the addition of MgATP to identify the changes that occur when this ε inhibition is removed. The maps generated show that, after exposure to MgATP, ATP synthase adopts a different conformation with a catalytic subunit changing conformation substantially and the ε C-terminal domain transitioning via an intermediate 'half-up' state to a condensed 'down' state. This work provides direct evidence for unique conformational states that occur in ATP synthase when ATP binding prevents the ε C-terminal domain from entering the inhibitory 'up' state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9346.map.gz emd_9346.map.gz | 286 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9346-v30.xml emd-9346-v30.xml emd-9346.xml emd-9346.xml | 8.5 KB 8.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9346.png emd_9346.png | 50.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9346 http://ftp.pdbj.org/pub/emdb/structures/EMD-9346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9346 | HTTPS FTP |
-Validation report
| Summary document |  emd_9346_validation.pdf.gz emd_9346_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9346_full_validation.pdf.gz emd_9346_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_9346_validation.xml.gz emd_9346_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9346 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9346 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9346 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9346 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9346.map.gz / Format: CCP4 / Size: 303.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9346.map.gz / Format: CCP4 / Size: 303.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli ATP synthase after incubation with ATP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
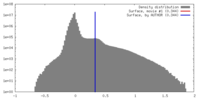
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. coli ATP synthase
| Entire | Name: E. coli ATP synthase |
|---|---|
| Components |
|
-Supramolecule #1: E. coli ATP synthase
| Supramolecule | Name: E. coli ATP synthase / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 532 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 5.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 45446 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)