[English] 日本語
 Yorodumi
Yorodumi- EMDB-9257: The structure of the Plasmodium falciparum 20S proteasome in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9257 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the Plasmodium falciparum 20S proteasome in complex with two PA28 activators. | |||||||||
 Map data Map data | Sharpened map used for structure refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | proteasome / protease / HYDROLASE / 11S subunit / hydrolyse activator / proteasome activator / complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationER-Phagosome pathway / Cross-presentation of soluble exogenous antigens (endosomes) / Proteasome assembly / Antigen processing: Ub, ATP-independent proteasomal degradation / proteasome activator complex / Orc1 removal from chromatin / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway / UCH proteinases ...ER-Phagosome pathway / Cross-presentation of soluble exogenous antigens (endosomes) / Proteasome assembly / Antigen processing: Ub, ATP-independent proteasomal degradation / proteasome activator complex / Orc1 removal from chromatin / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway / UCH proteinases / Ub-specific processing proteases / Neddylation / Antigen processing: Ubiquitination & Proteasome degradation / MAPK6/MAPK4 signaling / ABC-family proteins mediated transport / AUF1 (hnRNP D0) binds and destabilizes mRNA / Neutrophil degranulation / proteasome core complex / proteasomal ubiquitin-independent protein catabolic process / regulation of G1/S transition of mitotic cell cycle / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / endopeptidase activator activity / threonine-type endopeptidase activity / proteasome core complex, alpha-subunit complex / proteasomal protein catabolic process / regulation of proteasomal protein catabolic process / endopeptidase activity / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / hydrolase activity / nucleoplasm / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Hanssen E / Xie SC / Leis A / Metcalfe RD / Gillett DL / Tilley L / Griffin MDW | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2019 Journal: Nat Microbiol / Year: 2019Title: The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis. Authors: Stanley C Xie / Riley D Metcalfe / Eric Hanssen / Tuo Yang / David L Gillett / Andrew P Leis / Craig J Morton / Michael J Kuiper / Michael W Parker / Natalie J Spillman / Wilson Wong / ...Authors: Stanley C Xie / Riley D Metcalfe / Eric Hanssen / Tuo Yang / David L Gillett / Andrew P Leis / Craig J Morton / Michael J Kuiper / Michael W Parker / Natalie J Spillman / Wilson Wong / Christopher Tsu / Lawrence R Dick / Michael D W Griffin / Leann Tilley /   Abstract: The activity of the proteasome 20S catalytic core is regulated by protein complexes that bind to one or both ends. The PA28 regulator stimulates 20S proteasome peptidase activity in vitro, but its ...The activity of the proteasome 20S catalytic core is regulated by protein complexes that bind to one or both ends. The PA28 regulator stimulates 20S proteasome peptidase activity in vitro, but its role in vivo remains unclear. Here, we show that genetic deletion of the PA28 regulator from Plasmodium falciparum (Pf) renders malaria parasites more sensitive to the antimalarial drug dihydroartemisinin, indicating that PA28 may play a role in protection against proteotoxic stress. The crystal structure of PfPA28 reveals a bell-shaped molecule with an inner pore that has a strong segregation of charges. Small-angle X-ray scattering shows that disordered loops, which are not resolved in the crystal structure, extend from the PfPA28 heptamer and surround the pore. Using single particle cryo-electron microscopy, we solved the structure of Pf20S in complex with one and two regulatory PfPA28 caps at resolutions of 3.9 and 3.8 Å, respectively. PfPA28 binds Pf20S asymmetrically, strongly engaging subunits on only one side of the core. PfPA28 undergoes rigid body motions relative to Pf20S. Molecular dynamics simulations support conformational flexibility and a leaky interface. We propose lateral transfer of short peptides through the dynamic interface as a mechanism facilitating the release of proteasome degradation products. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9257.map.gz emd_9257.map.gz | 141.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9257-v30.xml emd-9257-v30.xml emd-9257.xml emd-9257.xml | 42 KB 42 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9257_fsc.xml emd_9257_fsc.xml | 12.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_9257.png emd_9257.png | 94.3 KB | ||
| Filedesc metadata |  emd-9257.cif.gz emd-9257.cif.gz | 9.8 KB | ||
| Others |  emd_9257_additional.map.gz emd_9257_additional.map.gz emd_9257_half_map_1.map.gz emd_9257_half_map_1.map.gz emd_9257_half_map_2.map.gz emd_9257_half_map_2.map.gz | 122.3 MB 122.3 MB 122.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9257 http://ftp.pdbj.org/pub/emdb/structures/EMD-9257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9257 | HTTPS FTP |
-Related structure data
| Related structure data |  6muvMC  9258C  9259C  6dfkC  6muwC  6muxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9257.map.gz / Format: CCP4 / Size: 155.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9257.map.gz / Format: CCP4 / Size: 155.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map used for structure refinement. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Initial unsharpened map.
| File | emd_9257_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Initial unsharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
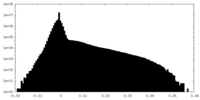
| Density Histograms |
-Half map: Plasmodium falciparum 20S proteasome
| File | emd_9257_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Plasmodium falciparum 20S proteasome | ||||||||||||
| Projections & Slices |
| ||||||||||||
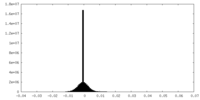
| Density Histograms |
-Half map: Plasmodium falciparum 20S proteasome
| File | emd_9257_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Plasmodium falciparum 20S proteasome | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 20S proteasome/PA28 complex.
+Supramolecule #1: 20S proteasome/PA28 complex.
+Supramolecule #2: Plasmodium falciparum 20S proteasome
+Supramolecule #3: 11S activator of the Plasmodium falciparum proteasome.
+Macromolecule #1: 20S proteasome alpha-1 subunit
+Macromolecule #2: 20S proteasome alpha-2 subunit
+Macromolecule #3: 20S proteasome alpha-3 subunit
+Macromolecule #4: 20S proteasome alpha-4 subunit
+Macromolecule #5: 20S proteasome alpha-5 subunit
+Macromolecule #6: 20S proteasome alpha-6 subunit
+Macromolecule #7: 20S proteasome alpha-7 subunit
+Macromolecule #8: 20S proteasome beta-1 subunit
+Macromolecule #9: 20S proteasome beta-2 subunit
+Macromolecule #10: 20S proteasome beta-3 subunit
+Macromolecule #11: 20S proteasome beta-4 subunit
+Macromolecule #12: 20S proteasome beta-5 subunit
+Macromolecule #13: 20S proteasome beta-6 subunit
+Macromolecule #14: 20S proteasome beta-7 subunit
+Macromolecule #15: Proteasome activator PA28
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Details: unspecified | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: wait time 0sec blot time 2sec blot force -1. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 5200 / Average exposure time: 10.0 sec. / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: 1.0 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: M / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 99.23 |
| Output model |  PDB-6muv: |
 Movie
Movie Controller
Controller














 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)