+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9242 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state) | |||||||||
 Map data Map data | Postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / translation | |||||||||
| Function / homology |  Function and homology information Function and homology informationL13a-mediated translational silencing of Ceruloplasmin expression / Translation initiation complex formation / Formation of a pool of free 40S subunits / Formation of the ternary complex, and subsequently, the 43S complex / Ribosomal scanning and start codon recognition / GTP hydrolysis and joining of the 60S ribosomal subunit / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Protein hydroxylation ...L13a-mediated translational silencing of Ceruloplasmin expression / Translation initiation complex formation / Formation of a pool of free 40S subunits / Formation of the ternary complex, and subsequently, the 43S complex / Ribosomal scanning and start codon recognition / GTP hydrolysis and joining of the 60S ribosomal subunit / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Protein hydroxylation / Translation initiation complex formation / Formation of the ternary complex, and subsequently, the 43S complex / Ribosomal scanning and start codon recognition / L13a-mediated translational silencing of Ceruloplasmin expression / SRP-dependent cotranslational protein targeting to membrane / Formation of a pool of free 40S subunits / GTP hydrolysis and joining of the 60S ribosomal subunit / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Major pathway of rRNA processing in the nucleolus and cytosol / SRP-dependent cotranslational protein targeting to membrane / L13a-mediated translational silencing of Ceruloplasmin expression / SRP-dependent cotranslational protein targeting to membrane / Major pathway of rRNA processing in the nucleolus and cytosol / Formation of a pool of free 40S subunits / GTP hydrolysis and joining of the 60S ribosomal subunit / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Translation initiation complex formation / Formation of the ternary complex, and subsequently, the 43S complex / Ribosomal scanning and start codon recognition / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / RHO GTPases Activate Formins / Major pathway of rRNA processing in the nucleolus and cytosol / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / SRP-dependent cotranslational protein targeting to membrane / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Separation of Sister Chromatids / TORC2 complex binding / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / Ribosomal scanning and start codon recognition / Translation initiation complex formation / SARS-CoV-1 modulates host translation machinery / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / Eukaryotic Translation Termination / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / ubiquitin ligase inhibitor activity / Viral mRNA Translation / 90S preribosome / positive regulation of signal transduction by p53 class mediator / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / phagocytic cup / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / protein-RNA complex assembly / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rough endoplasmic reticulum / ribosomal small subunit export from nucleus / translation regulator activity / translation initiation factor binding / gastrulation / MDM2/MDM4 family protein binding / stress granule assembly / cytosolic ribosome / class I DNA-(apurinic or apyrimidinic site) endonuclease activity / DNA-(apurinic or apyrimidinic site) lyase / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / erythrocyte differentiation / positive regulation of apoptotic signaling pathway / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / maintenance of translational fidelity / mRNA 5'-UTR binding / spindle / Regulation of expression of SLITs and ROBOs / cytoplasmic ribonucleoprotein granule / rRNA processing / positive regulation of canonical Wnt signaling pathway / rhythmic process / antimicrobial humoral immune response mediated by antimicrobial peptide / regulation of translation / large ribosomal subunit / ribosome biogenesis / ribosome binding / ribosomal small subunit biogenesis Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Brown A / Baird MR / Yip MCJ / Murray J / Shao S | |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: Structures of translationally inactive mammalian ribosomes. Authors: Alan Brown / Matthew R Baird / Matthew Cj Yip / Jason Murray / Sichen Shao /   Abstract: The cellular levels and activities of ribosomes directly regulate gene expression during numerous physiological processes. The mechanisms that globally repress translation are incompletely understood. ...The cellular levels and activities of ribosomes directly regulate gene expression during numerous physiological processes. The mechanisms that globally repress translation are incompletely understood. Here, we use electron cryomicroscopy to analyze inactive ribosomes isolated from mammalian reticulocytes, the penultimate stage of red blood cell differentiation. We identify two types of ribosomes that are translationally repressed by protein interactions. The first comprises ribosomes sequestered with elongation factor 2 (eEF2) by SERPINE mRNA binding protein 1 (SERBP1) occupying the ribosomal mRNA entrance channel. The second type are translationally repressed by a novel ribosome-binding protein, interferon-related developmental regulator 2 (IFRD2), which spans the P and E sites and inserts a C-terminal helix into the mRNA exit channel to preclude translation. IFRD2 binds ribosomes with a tRNA occupying a noncanonical binding site, the 'Z site', on the ribosome. These structures provide functional insights into how ribosomal interactions may suppress translation to regulate gene expression. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9242.map.gz emd_9242.map.gz | 12.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9242-v30.xml emd-9242-v30.xml emd-9242.xml emd-9242.xml | 104.5 KB 104.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9242_fsc.xml emd_9242_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9242.png emd_9242.png | 172 KB | ||
| Filedesc metadata |  emd-9242.cif.gz emd-9242.cif.gz | 22.4 KB | ||
| Others |  emd_9242_additional.map.gz emd_9242_additional.map.gz emd_9242_half_map_1.map.gz emd_9242_half_map_1.map.gz emd_9242_half_map_2.map.gz emd_9242_half_map_2.map.gz | 214.2 MB 214.5 MB 214.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9242 http://ftp.pdbj.org/pub/emdb/structures/EMD-9242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9242 | HTTPS FTP |
-Related structure data
| Related structure data |  6mteMC  9234C  9235C  9236C  9237C  9239C  9240C  9241C  6mtbC  6mtcC  6mtdC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9242.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9242.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
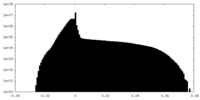
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
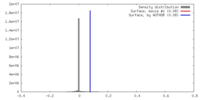
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Pre-postprocessed map
| File | emd_9242_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pre-postprocessed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_9242_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_9242_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state)
+Supramolecule #1: Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state)
+Macromolecule #1: 28S rRNA
+Macromolecule #2: 5S rRNA
+Macromolecule #3: 5.8S rRNA
+Macromolecule #50: 18S rRNA
+Macromolecule #4: uL2
+Macromolecule #5: uL3
+Macromolecule #6: uL4
+Macromolecule #7: uL18
+Macromolecule #8: eL6
+Macromolecule #9: uL30
+Macromolecule #10: eL8
+Macromolecule #11: uL6
+Macromolecule #12: uL16
+Macromolecule #13: uL5
+Macromolecule #14: eL13
+Macromolecule #15: eL14
+Macromolecule #16: eL15
+Macromolecule #17: uL13
+Macromolecule #18: uL22
+Macromolecule #19: eL18
+Macromolecule #20: eL19
+Macromolecule #21: eL20
+Macromolecule #22: eL21
+Macromolecule #23: eL22
+Macromolecule #24: uL14
+Macromolecule #25: eL24
+Macromolecule #26: uL23
+Macromolecule #27: uL24
+Macromolecule #28: eL27
+Macromolecule #29: uL15
+Macromolecule #30: eL29
+Macromolecule #31: eL30
+Macromolecule #32: eL31
+Macromolecule #33: eL32
+Macromolecule #34: eL33
+Macromolecule #35: eL34
+Macromolecule #36: uL29
+Macromolecule #37: eL36
+Macromolecule #38: eL37
+Macromolecule #39: eL38
+Macromolecule #40: eL39
+Macromolecule #41: eL40
+Macromolecule #42: eL41
+Macromolecule #43: eL42
+Macromolecule #44: eL43
+Macromolecule #45: eL28
+Macromolecule #46: uL10
+Macromolecule #47: uL11
+Macromolecule #48: eEF2
+Macromolecule #49: SERBP1
+Macromolecule #51: uS2
+Macromolecule #52: eS1
+Macromolecule #53: uS5
+Macromolecule #54: uS3
+Macromolecule #55: eS4
+Macromolecule #56: uS7
+Macromolecule #57: eS6
+Macromolecule #58: eS7
+Macromolecule #59: eS8
+Macromolecule #60: uS4
+Macromolecule #61: eS10
+Macromolecule #62: uS17
+Macromolecule #63: eS12
+Macromolecule #64: uS15
+Macromolecule #65: uS11
+Macromolecule #66: uS19
+Macromolecule #67: uS9
+Macromolecule #68: eS17
+Macromolecule #69: uS13
+Macromolecule #70: eS19
+Macromolecule #71: uS10
+Macromolecule #72: eS21
+Macromolecule #73: uS8
+Macromolecule #74: uS12
+Macromolecule #75: eS24
+Macromolecule #76: eS25
+Macromolecule #77: eS26
+Macromolecule #78: eS27
+Macromolecule #79: eS28
+Macromolecule #80: uS14
+Macromolecule #81: eS30
+Macromolecule #82: eS31
+Macromolecule #83: RACK1
+Macromolecule #84: MAGNESIUM ION
+Macromolecule #85: ZINC ION
+Macromolecule #86: GUANOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Frames/image: 1-17 / Average exposure time: 1.1 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 104478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)