[English] 日本語
 Yorodumi
Yorodumi- EMDB-4129: Structure of the mammalian ribosome with P- and E-site tRNAs and ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4129 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the mammalian ribosome with P- and E-site tRNAs and an unoccupied A site | |||||||||
 Map data Map data | Postprocessed, sharpened map. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Shao S / Murray J / Brown A / Taunton J / Ramakrishnan V / Hegde RS | |||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Authors: Sichen Shao / Jason Murray / Alan Brown / Jack Taunton / V Ramakrishnan / Ramanujan S Hegde /   Abstract: In eukaryotes, accurate protein synthesis relies on a family of translational GTPases that pair with specific decoding factors to decipher the mRNA code on ribosomes. We present structures of the ...In eukaryotes, accurate protein synthesis relies on a family of translational GTPases that pair with specific decoding factors to decipher the mRNA code on ribosomes. We present structures of the mammalian ribosome engaged with decoding factor⋅GTPase complexes representing intermediates of translation elongation (aminoacyl-tRNA⋅eEF1A), termination (eRF1⋅eRF3), and ribosome rescue (Pelota⋅Hbs1l). Comparative analyses reveal that each decoding factor exploits the plasticity of the ribosomal decoding center to differentially remodel ribosomal proteins and rRNA. This leads to varying degrees of large-scale ribosome movements and implies distinct mechanisms for communicating information from the decoding center to each GTPase. Additional structural snapshots of the translation termination pathway reveal the conformational changes that choreograph the accommodation of decoding factors into the peptidyl transferase center. Our results provide a structural framework for how different states of the mammalian ribosome are selectively recognized by the appropriate decoding factor⋅GTPase complex to ensure translational fidelity. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Decoding mammalian ribosome-mRNA states by translational GTPase complexes Authors: Shao S / Murray J / Brown A / Taunton J / Ramakrishnan V / Hegde RS | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4129.map.gz emd_4129.map.gz | 24.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4129-v30.xml emd-4129-v30.xml emd-4129.xml emd-4129.xml | 28.6 KB 28.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4129_fsc.xml emd_4129_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4129.png emd_4129.png | 196.8 KB | ||
| Others |  emd_4129_half_map_1.map.gz emd_4129_half_map_1.map.gz emd_4129_half_map_2.map.gz emd_4129_half_map_2.map.gz | 311.5 MB 313 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4129 http://ftp.pdbj.org/pub/emdb/structures/EMD-4129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4129 | HTTPS FTP |
-Related structure data
| Related structure data |  4130C  4131C  4132C  4133C  4134C  4135C  4136C  4137C  5lzsC  5lztC  5lzuC  5lzvC  5lzwC  5lzxC  5lzyC  5lzzC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4129.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4129.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed, sharpened map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
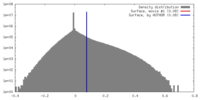
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1.
| File | emd_4129_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
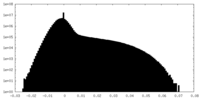
| Density Histograms |
-Half map: Half map 2.
| File | emd_4129_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Affinity-purified 80S ribosome-nascent chain complex.
| Entire | Name: Affinity-purified 80S ribosome-nascent chain complex. |
|---|---|
| Components |
|
-Supramolecule #1: Affinity-purified 80S ribosome-nascent chain complex.
| Supramolecule | Name: Affinity-purified 80S ribosome-nascent chain complex. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#85 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III Details: 3 ul aliquots were applied to the grid and incubated for 30 s, before blotting for 3 s to remove excess solution.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number real images: 1643 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 134615 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 69000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Overall B value: 51.6 |
|---|
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)