[English] 日本語
 Yorodumi
Yorodumi- EMDB-8956: Structure of AtTPC1(DDE) reconstituted in saposin A with cat06 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8956 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of AtTPC1(DDE) reconstituted in saposin A with cat06 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Two-pore channel / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of jasmonic acid biosynthetic process / seed germination / regulation of stomatal movement / plant-type vacuole / vacuole / vacuolar membrane / monoatomic ion channel complex / voltage-gated calcium channel activity / calcium-mediated signaling / calcium ion transport ...regulation of jasmonic acid biosynthetic process / seed germination / regulation of stomatal movement / plant-type vacuole / vacuole / vacuolar membrane / monoatomic ion channel complex / voltage-gated calcium channel activity / calcium-mediated signaling / calcium ion transport / calcium ion binding / Golgi apparatus / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
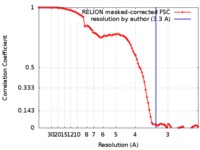
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Green EM / Kintzer AF / Stroud RM / Cheng Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Structural basis for activation of voltage sensor domains in an ion channel TPC1. Authors: Alexander F Kintzer / Evan M Green / Pawel K Dominik / Michael Bridges / Jean-Paul Armache / Dawid Deneka / Sangwoo S Kim / Wayne Hubbell / Anthony A Kossiakoff / Yifan Cheng / Robert M Stroud /  Abstract: Voltage-sensing domains (VSDs) couple changes in transmembrane electrical potential to conformational changes that regulate ion conductance through a central channel. Positively charged amino acids ...Voltage-sensing domains (VSDs) couple changes in transmembrane electrical potential to conformational changes that regulate ion conductance through a central channel. Positively charged amino acids inside each sensor cooperatively respond to changes in voltage. Our previous structure of a TPC1 channel captured an example of a resting-state VSD in an intact ion channel. To generate an activated-state VSD in the same channel we removed the luminal inhibitory Ca-binding site (Ca), which shifts voltage-dependent opening to more negative voltage and activation at 0 mV. Cryo-EM reveals two coexisting structures of the VSD, an intermediate state 1 that partially closes access to the cytoplasmic side but remains occluded on the luminal side and an intermediate activated state 2 in which the cytoplasmic solvent access to the gating charges closes, while luminal access partially opens. Activation can be thought of as moving a hydrophobic insulating region of the VSD from the external side to an alternate grouping on the internal side. This effectively moves the gating charges from the inside potential to that of the outside. Activation also requires binding of Ca to a cytoplasmic site (Ca). An X-ray structure with Ca removed and a near-atomic resolution cryo-EM structure with Ca removed define how dramatic conformational changes in the cytoplasmic domains may communicate with the VSD during activation. Together four structures provide a basis for understanding the voltage-dependent transition from resting to activated state, the tuning of VSD by thermodynamic stability, and this channel's requirement of cytoplasmic Ca ions for activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8956.map.gz emd_8956.map.gz | 60.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8956-v30.xml emd-8956-v30.xml emd-8956.xml emd-8956.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8956_fsc.xml emd_8956_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_8956.png emd_8956.png | 93.6 KB | ||
| Filedesc metadata |  emd-8956.cif.gz emd-8956.cif.gz | 6.7 KB | ||
| Others |  emd_8956_half_map_1.map.gz emd_8956_half_map_1.map.gz emd_8956_half_map_2.map.gz emd_8956_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8956 http://ftp.pdbj.org/pub/emdb/structures/EMD-8956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8956 | HTTPS FTP |
-Related structure data
| Related structure data |  6e1kMC  8957C  8958C  8960C  6cx0C  6e1mC  6e1nC  6e1pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8956.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8956.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2156 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
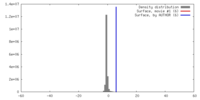
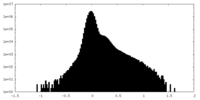
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_8956_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
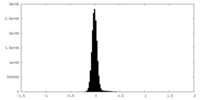
| Density Histograms |
-Half map: #1
| File | emd_8956_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
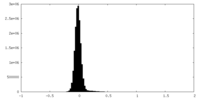
| Density Histograms |
- Sample components
Sample components
-Entire : AtTPC1(DDE) reconstituted in saposin A
| Entire | Name: AtTPC1(DDE) reconstituted in saposin A |
|---|---|
| Components |
|
-Supramolecule #1: AtTPC1(DDE) reconstituted in saposin A
| Supramolecule | Name: AtTPC1(DDE) reconstituted in saposin A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Two pore calcium channel protein 1
| Macromolecule | Name: Two pore calcium channel protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.547203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGGGTDRVR R(SEP)EAI(TPO)HG(TPO)P FQKAAALVDL AEDGIGLPVE ILDQSSFGES ARYYFIFTRL DLIWSLNY F ALLFLNFFEQ PLWCEKNPKP SCKDRDYYYL GELPYLTNAE SIIYEVITLA ILLVHTFFPI SYEGSRIFWT SRLNLVKVA CVVILFVDVL ...String: MGGGGTDRVR R(SEP)EAI(TPO)HG(TPO)P FQKAAALVDL AEDGIGLPVE ILDQSSFGES ARYYFIFTRL DLIWSLNY F ALLFLNFFEQ PLWCEKNPKP SCKDRDYYYL GELPYLTNAE SIIYEVITLA ILLVHTFFPI SYEGSRIFWT SRLNLVKVA CVVILFVDVL VDFLYLSPLA FDFLPFRIAP YVRVIIFILS IRELRDTLVL LSGMLGTYLN ILALWMLFLL FASWIAFVMF ENTQQGLTV FTSYGATLYQ MFILFTTSNN PDVWIPAYKS SRWSSVFFVL YVLIGVYFVT NLILAVVYDS FKEQLAKQVS G MDQMKRRM LEKAFGLIDS DKNGEIDKNQ CIKLFEQLTN YRTLPKISKE EFGLIFDELD DTRDFKINKD EFADLCQAIA LR FQKEEVP SLFEHFPQIY HSALSQQLRA FVRSPNFGYA ISFILIINFI AVVVETTLNI EESSAQKPWQ VAEFVFGWIY VLE MALKIY TYGFENYWRE GANRFDFLVT WVIVIGETAT FITPDENTFF SNGQWIRYLL LARMLRLIRL LMNVQRYRAF IATF ITLIP SLMPYLGTIF CVLCIYCSIG VQVFGGLVNA GNKKLFETEL AEDDYLLFNF NDYPNGMVTL FNLLVMGNWQ VWMES YKDL TGTWWSITYF VSFYVITILL LLNLVVAFVL EAFFTELDLE EEEKCQGQDS QEKRNRRRSA GSKSRSQRVD TLLHHM LGD ELSKPECSTS DTLVPR UniProtKB: Two pore calcium channel protein 1 |
-Macromolecule #2: cat06 light chain
| Macromolecule | Name: cat06 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.171703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QSSSSLITFG QGTKVEIKRT VAAPSVFIFP PSDSQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QSSSSLITFG QGTKVEIKRT VAAPSVFIFP PSDSQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: cat06 heavy chain
| Macromolecule | Name: cat06 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.786658 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFNVY SYSIHWVRQA PGKGLEWVAS ISSYYSSTSY ADSVKGRFTI SADTSKNTAY LQMNSLRAE DTAVYYCARS YWYWRTSTLG GIDYWGQGTL VTVFNQIKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: EVQLVESGGG LVQPGGSLRL SCAASGFNVY SYSIHWVRQA PGKGLEWVAS ISSYYSSTSY ADSVKGRFTI SADTSKNTAY LQMNSLRAE DTAVYYCARS YWYWRTSTLG GIDYWGQGTL VTVFNQIKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDK THT |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 7 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 5 / Number of copies: 14 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.3 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 20 K / Instrument: FEI VITROBOT MARK III |
| Details | Sample reconstituted into saposin A. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 2-60 / Number grids imaged: 2 / Number real images: 3408 / Average exposure time: 0.2 sec. / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 41132 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 31000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6e1k: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)