[English] 日本語
 Yorodumi
Yorodumi- EMDB-8761: CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8761 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degrees Celsius, molar ratio 1:3, full particle) | |||||||||
 Map data Map data | Inner density masked out. The map has been sharpened. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / antibody / VIRUS-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysis of host organelle involved in viral entry into host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...lysis of host organelle involved in viral entry into host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Human rhinovirus 14 / Human rhinovirus 14 /   Human rhinovirus B14 Human rhinovirus B14 | |||||||||
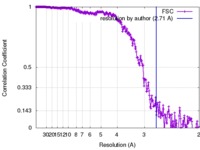
| Method | single particle reconstruction / cryo EM / Resolution: 2.71 Å | |||||||||
 Authors Authors | Liu Y / Dong Y / Rossmann MG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Antibody-induced uncoating of human rhinovirus B14. Authors: Yangchao Dong / Yue Liu / Wen Jiang / Thomas J Smith / Zhikai Xu / Michael G Rossmann /   Abstract: Rhinoviruses (RVs) are the major causes of common colds in humans. They have a nonenveloped, icosahedral capsid surrounding a positive-strand RNA genome. Here we report that the antigen-binding (Fab) ...Rhinoviruses (RVs) are the major causes of common colds in humans. They have a nonenveloped, icosahedral capsid surrounding a positive-strand RNA genome. Here we report that the antigen-binding (Fab) fragment of a neutralizing antibody (C5) can trigger genome release from RV-B14 to form emptied particles and neutralize virus infection. Using cryo-electron microscopy, structures of the C5 Fab in complex with the full and emptied particles have been determined at 2.3 Å and 3.0 Å resolution, respectively. Each of the 60 Fab molecules binds primarily to a region on viral protein 3 (VP3). Binding of the C5 Fabs to RV-B14 results in significant conformational changes around holes in the capsid through which the viral RNA might exit. These results are so far the highest resolution view of an antibody-virus complex and elucidate a mechanism whereby antibodies neutralize RVs and related viruses by inducing virus uncoating. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8761.map.gz emd_8761.map.gz | 1.1 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8761-v30.xml emd-8761-v30.xml emd-8761.xml emd-8761.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8761_fsc.xml emd_8761_fsc.xml | 23.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_8761.png emd_8761.png | 250.8 KB | ||
| Filedesc metadata |  emd-8761.cif.gz emd-8761.cif.gz | 6.8 KB | ||
| Others |  emd_8761_half_map_1.map.gz emd_8761_half_map_1.map.gz emd_8761_half_map_2.map.gz emd_8761_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8761 http://ftp.pdbj.org/pub/emdb/structures/EMD-8761 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8761 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8761 | HTTPS FTP |
-Related structure data
| Related structure data |  5w3lMC  8754C  8762C  8763C  5w3eC  5w3mC  5w3oC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8761.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8761.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Inner density masked out. The map has been sharpened. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Unmasked, unsharpened half map #1
| File | emd_8761_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked, unsharpened half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unmasked, unsharpened half map #2
| File | emd_8761_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked, unsharpened half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human rhinovirus B14
| Entire | Name:  Human rhinovirus B14 Human rhinovirus B14 |
|---|---|
| Components |
|
-Supramolecule #1: Human rhinovirus B14
| Supramolecule | Name: Human rhinovirus B14 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 / Details: Viruses were grown in HeLa-H1 cells. / NCBI-ID: 12131 / Sci species name: Human rhinovirus B14 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: viral protein 1
| Macromolecule | Name: viral protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human rhinovirus 14 Human rhinovirus 14 |
| Molecular weight | Theoretical: 32.560549 KDa |
| Sequence | String: GLGDELEEVI VEKTKQTVAS ISSGPKHTQK VPILTANETG ATMPVLPSDS IETRTTYMHF NGSETDVECF LGRAACVHVT EIQNKDATG IDNHREAKLF NDWKINLSSL VQLRKKLELF TYVRFDSEYT ILATASQPDS ANYSSNLVVQ AMYVPPGAPN P KEWDDYTW ...String: GLGDELEEVI VEKTKQTVAS ISSGPKHTQK VPILTANETG ATMPVLPSDS IETRTTYMHF NGSETDVECF LGRAACVHVT EIQNKDATG IDNHREAKLF NDWKINLSSL VQLRKKLELF TYVRFDSEYT ILATASQPDS ANYSSNLVVQ AMYVPPGAPN P KEWDDYTW QSASNPSVFF KVGDTSRFSV PYVGLASAYN CFYDGYSHDD AETQYGITVL NHMGSMAFRI VNEHDEHKTL VK IRVYHRA KHVEAWIPRA PRALPYTSIG RTNYPKNTEP VIKKRKGDIK SY UniProtKB: Genome polyprotein |
-Macromolecule #2: viral protein 3
| Macromolecule | Name: viral protein 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human rhinovirus 14 Human rhinovirus 14 |
| Molecular weight | Theoretical: 26.236754 KDa |
| Sequence | String: GLPTTTLPGS GQFLTTDDRQ SPSALPNYEP TPRIHIPGKV HNLLEIIQVD TLIPMNNTHT KDEVNSYLIP LNANRQNEQV FGTNLFIGD GVFKTTLLGE IVQYYTHWSG SLRFSLMYTG PALSSAKLIL AYTPPGARGP QDRREAMLGT HVVWDIGLQS T IVMTIPWT ...String: GLPTTTLPGS GQFLTTDDRQ SPSALPNYEP TPRIHIPGKV HNLLEIIQVD TLIPMNNTHT KDEVNSYLIP LNANRQNEQV FGTNLFIGD GVFKTTLLGE IVQYYTHWSG SLRFSLMYTG PALSSAKLIL AYTPPGARGP QDRREAMLGT HVVWDIGLQS T IVMTIPWT SGVQFRYTDP DTYTSAGFLS CWYQTSLILP PETTGQVYLL SFISACPDFK LRLMKDTQTI SQTVALTE UniProtKB: Genome polyprotein |
-Macromolecule #3: viral protein 2
| Macromolecule | Name: viral protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human rhinovirus 14 Human rhinovirus 14 |
| Molecular weight | Theoretical: 28.501361 KDa |
| Sequence | String: SPNVEACGYS DRVQQITLGN STITTQEAAN AVVCYAEWPE YLPDVDASDV NKTSKPDTSV CRFYTLDSKT WTTGSKGWCW KLPDALKDM GVFGQNMFFH SLGRSGYTVH VQCNATKFHS GCLLVVVIPE HQLASHEGGN VSVKYTFTHP GERGIDLSSA N EVGGPVKD ...String: SPNVEACGYS DRVQQITLGN STITTQEAAN AVVCYAEWPE YLPDVDASDV NKTSKPDTSV CRFYTLDSKT WTTGSKGWCW KLPDALKDM GVFGQNMFFH SLGRSGYTVH VQCNATKFHS GCLLVVVIPE HQLASHEGGN VSVKYTFTHP GERGIDLSSA N EVGGPVKD VIYNMNGTLL GNLLIFPHQF INLRTNNTAT IVIPYINSVP IDSMTRHNNV SLMVIPIAPL TVPTGATPSL PI TVTIAPM CTEFSGIRSK SIVPQ UniProtKB: Genome polyprotein |
-Macromolecule #4: viral protein 4
| Macromolecule | Name: viral protein 4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human rhinovirus 14 Human rhinovirus 14 |
| Molecular weight | Theoretical: 7.183863 KDa |
| Sequence | String: GAQVSTQKSG SHENQNILTN GSNQTFTVIN YYKDAASTSS AGQSLSMDPS KFTEPVKDLM LKGAPALN UniProtKB: Genome polyprotein |
-Macromolecule #5: C5 antibody variable heavy domain
| Macromolecule | Name: C5 antibody variable heavy domain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.989335 KDa |
| Sequence | String: AVQLAESGPA LVAPSQALSI TCTVAGFSLT AYGVAWVRQP PGAGLEWLGA IWAAGATDYN AALKSRASIA KDNSKSQVFL AMASLATAD TAAYYCAREW DAYGDYWGQG TTVTVSA |
-Macromolecule #6: C5 antibody variable light domain
| Macromolecule | Name: C5 antibody variable light domain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.897161 KDa |
| Sequence | String: DIVLTQSPAA LSAAAGATVA ATCRASGNIH NALAWYQQKA GKSPQLLVYA AAALAAGVPS RFSGSGSGTA YALAINSLAA DDFGAYYCQ HFWSTPYTFG GGTKLEIK |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 166 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 20 mM Tris, 120 mM sodium chloride, 1 mM EDTA, pH 8.0 |
|---|---|
| Grid | Model: Ted Pella, Ultrathin lacey carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 4-49 / Number grids imaged: 1 / Number real images: 537 / Average exposure time: 6.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | A combination of the following approaches was used: (1) model rebuilding using Coot, (2) real space refinement using Phenix, (3) reciprocal space refinment using Phenix (as in standard crystallographic refinement). |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coeffcient |
| Output model |  PDB-5w3l: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)