+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-8672 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | |||||||||
 マップデータ マップデータ | The final map of RP corrected with a B-factor of -50 | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | p28 / 26S proteasome / regulatory particle / 19S / gankyrin / HYDROLASE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; オメガペプチターゼ / integrator complex / proteasome accessory complex / meiosis I ...Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; オメガペプチターゼ / integrator complex / proteasome accessory complex / meiosis I / proteasome regulatory particle / cytosolic proteasome complex / positive regulation of proteasomal protein catabolic process / proteasome-activating activity / proteasome regulatory particle, lid subcomplex / proteasome regulatory particle, base subcomplex / metal-dependent deubiquitinase activity / protein K63-linked deubiquitination / negative regulation of programmed cell death / Regulation of ornithine decarboxylase (ODC) / Proteasome assembly / Cross-presentation of soluble exogenous antigens (endosomes) / Homologous DNA Pairing and Strand Exchange / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / Resolution of D-loop Structures through Holliday Junction Intermediates / Somitogenesis / K63-linked deubiquitinase activity / Impaired BRCA2 binding to RAD51 / proteasome binding / transcription factor binding / regulation of protein catabolic process / proteasome storage granule / Presynaptic phase of homologous DNA pairing and strand exchange / general transcription initiation factor binding / blastocyst development / polyubiquitin modification-dependent protein binding / positive regulation of RNA polymerase II transcription preinitiation complex assembly / protein deubiquitination / endopeptidase activator activity / mRNA export from nucleus / proteasome assembly / SARS-CoV-1 targets host intracellular signalling and regulatory pathways / enzyme regulator activity / regulation of proteasomal protein catabolic process / ERAD pathway / inclusion body / proteasome complex / TBP-class protein binding / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Asymmetric localization of PCP proteins / Ubiquitin-dependent degradation of Cyclin D / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / TNFR2 non-canonical NF-kB pathway / AUF1 (hnRNP D0) binds and destabilizes mRNA / stem cell differentiation / Vpu mediated degradation of CD4 / Assembly of the pre-replicative complex / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Degradation of DVL / Dectin-1 mediated noncanonical NF-kB signaling / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Degradation of AXIN / P-body / Hh mutants are degraded by ERAD / Activation of NF-kappaB in B cells / Degradation of GLI1 by the proteasome / G2/M Checkpoints / Hedgehog ligand biogenesis / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / Defective CFTR causes cystic fibrosis / Autodegradation of the E3 ubiquitin ligase COP1 / Negative regulation of NOTCH4 signaling / Regulation of RUNX3 expression and activity / Vif-mediated degradation of APOBEC3G / Hedgehog 'on' state / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / MAPK6/MAPK4 signaling / double-strand break repair via homologous recombination / Degradation of beta-catenin by the destruction complex / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / HDR through Homologous Recombination (HRR) / ABC-family proteins mediated transport / double-strand break repair via nonhomologous end joining / CDK-mediated phosphorylation and removal of Cdc6 / CLEC7A (Dectin-1) signaling / SCF(Skp2)-mediated degradation of p27/p21 / FCERI mediated NF-kB activation / Regulation of expression of SLITs and ROBOs / Metalloprotease DUBs / Regulation of PTEN stability and activity 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.5 Å | |||||||||
 データ登録者 データ登録者 | Lu Y / Wu J | |||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2017 ジャーナル: Mol Cell / 年: 2017タイトル: Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle. 著者: Ying Lu / Jiayi Wu / Yuanchen Dong / Shuobing Chen / Shuangwu Sun / Yong-Bei Ma / Qi Ouyang / Daniel Finley / Marc W Kirschner / Youdong Mao /   要旨: The proteasome holoenzyme is activated by its regulatory particle (RP) consisting of two subcomplexes, the lid and the base. A key event in base assembly is the formation of a heterohexameric ring of ...The proteasome holoenzyme is activated by its regulatory particle (RP) consisting of two subcomplexes, the lid and the base. A key event in base assembly is the formation of a heterohexameric ring of AAA-ATPases, which is guided by at least four RP assembly chaperones in mammals: PAAF1, p28/gankyrin, p27/PSMD9, and S5b. Using cryogenic electron microscopy, we analyzed the non-AAA structure of the p28-bound human RP at 4.5 Å resolution and determined seven distinct conformations of the Rpn1-p28-AAA subcomplex within the p28-bound RP at subnanometer resolutions. Remarkably, the p28-bound AAA ring does not form a channel in the free RP and spontaneously samples multiple "open" and "closed" topologies at the Rpt2-Rpt6 and Rpt3-Rpt4 interfaces. Our analysis suggests that p28 assists the proteolytic core particle to select a specific conformation of the ATPase ring for RP engagement and is released in a shoehorn-like fashion in the last step of the chaperone-mediated proteasome assembly. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_8672.map.gz emd_8672.map.gz | 58 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-8672-v30.xml emd-8672-v30.xml emd-8672.xml emd-8672.xml | 32.6 KB 32.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_8672.png emd_8672.png | 76.6 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8672 http://ftp.pdbj.org/pub/emdb/structures/EMD-8672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8672 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  5vgzMC  8674C  8675C  8676C  8677C  8678C  8679C  8680C  8681C  8682C  8683C  8684C  5vhfC  5vhhC  5vhiC  5vhjC  5vhmC  5vhnC  5vhoC  5vhpC  5vhqC  5vhrC  5vhsC C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-10091 (タイトル: Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle EMPIAR-10091 (タイトル: Conformational Landscape of the p28-Bound Human Proteasome Regulatory ParticleData size: 70.0 Data #1: Classified single-particle datasets for multiple conformations of p28-bound human regulatory complex [picked particles - multiframe - processed]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_8672.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_8672.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | The final map of RP corrected with a B-factor of -50 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
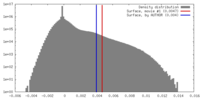
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Proteasome regulatory particle
+超分子 #1: Proteasome regulatory particle
+分子 #1: 26S proteasome regulatory subunit 7
+分子 #2: 26S proteasome regulatory subunit 4
+分子 #3: 26S proteasome regulatory subunit 8
+分子 #4: 26S proteasome regulatory subunit 6B
+分子 #5: 26S proteasome regulatory subunit 10B
+分子 #6: 26S proteasome regulatory subunit 6A
+分子 #7: 26S proteasome non-ATPase regulatory subunit 1
+分子 #8: 26S proteasome non-ATPase regulatory subunit 3
+分子 #9: 26S proteasome non-ATPase regulatory subunit 12
+分子 #10: 26S proteasome non-ATPase regulatory subunit 11
+分子 #11: 26S proteasome non-ATPase regulatory subunit 6
+分子 #12: 26S proteasome non-ATPase regulatory subunit 7
+分子 #13: 26S proteasome non-ATPase regulatory subunit 13
+分子 #14: 26S proteasome non-ATPase regulatory subunit 4
+分子 #15: 26S proteasome non-ATPase regulatory subunit 14
+分子 #16: 26S proteasome non-ATPase regulatory subunit 8
+分子 #17: 26S proteasome complex subunit SEM1
+分子 #18: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TECNAI ARCTICA |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: INSILICO MODEL |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 4.5 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 117471 |
| 初期 角度割当 | タイプ: ANGULAR RECONSTITUTION |
| 最終 角度割当 | タイプ: PROJECTION MATCHING |
 ムービー
ムービー コントローラー
コントローラー




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)