[English] 日本語
 Yorodumi
Yorodumi- EMDB-8683: Conformational Landscape of the p28-Bound Human Proteasome Regula... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8683 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | |||||||||
 Map data Map data | Final Map of Rpn1-p28-AAA subcomplex in the TA7 state, corrected with a B-factor of -400 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | p28 / 26S proteasome / regulatory particle / 19S / gankyrin / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationproteasome regulatory particle assembly / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / positive regulation of cyclin-dependent protein serine/threonine kinase activity / proteasome accessory complex / proteasome regulatory particle / cytosolic proteasome complex / positive regulation of proteasomal protein catabolic process ...proteasome regulatory particle assembly / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / positive regulation of cyclin-dependent protein serine/threonine kinase activity / proteasome accessory complex / proteasome regulatory particle / cytosolic proteasome complex / positive regulation of proteasomal protein catabolic process / proteasome-activating activity / proteasome regulatory particle, base subcomplex / negative regulation of programmed cell death / Regulation of ornithine decarboxylase (ODC) / Proteasome assembly / Cross-presentation of soluble exogenous antigens (endosomes) / Somitogenesis / : / negative regulation of release of cytochrome c from mitochondria / transcription factor binding / regulation of protein catabolic process / negative regulation of DNA damage response, signal transduction by p53 class mediator / proteasome storage granule / general transcription initiation factor binding / positive regulation of RNA polymerase II transcription preinitiation complex assembly / blastocyst development / enzyme regulator activity / negative regulation of MAPK cascade / SARS-CoV-1 targets host intracellular signalling and regulatory pathways / ERAD pathway / inclusion body / proteasome complex / TBP-class protein binding / positive regulation of protein ubiquitination / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Asymmetric localization of PCP proteins / Ubiquitin-dependent degradation of Cyclin D / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / TNFR2 non-canonical NF-kB pathway / AUF1 (hnRNP D0) binds and destabilizes mRNA / Assembly of the pre-replicative complex / Vpu mediated degradation of CD4 / P-body / Degradation of DVL / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Dectin-1 mediated noncanonical NF-kB signaling / Degradation of AXIN / Hh mutants are degraded by ERAD / Activation of NF-kappaB in B cells / G2/M Checkpoints / Degradation of GLI1 by the proteasome / Hedgehog ligand biogenesis / Defective CFTR causes cystic fibrosis / Autodegradation of the E3 ubiquitin ligase COP1 / Regulation of RUNX3 expression and activity / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / Negative regulation of NOTCH4 signaling / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Hedgehog 'on' state / Vif-mediated degradation of APOBEC3G / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / MAPK6/MAPK4 signaling / Degradation of beta-catenin by the destruction complex / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / ABC-family proteins mediated transport / CDK-mediated phosphorylation and removal of Cdc6 / CLEC7A (Dectin-1) signaling / SCF(Skp2)-mediated degradation of p27/p21 / FCERI mediated NF-kB activation / Regulation of expression of SLITs and ROBOs / Regulation of PTEN stability and activity / Interleukin-1 signaling / cytoplasmic ribonucleoprotein granule / Orc1 removal from chromatin / Regulation of RAS by GAPs / Regulation of RUNX2 expression and activity / osteoblast differentiation / The role of GTSE1 in G2/M progression after G2 checkpoint / Separation of Sister Chromatids / UCH proteinases / KEAP1-NFE2L2 pathway / Downstream TCR signaling / Antigen processing: Ubiquitination & Proteasome degradation / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / RUNX1 regulates transcription of genes involved in differentiation of HSCs / microtubule cytoskeleton / Neddylation / ER-Phagosome pathway / positive regulation of cell growth / cytoplasmic vesicle / secretory granule lumen / blood microparticle / protein-macromolecule adaptor activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.7 Å | |||||||||
 Authors Authors | Lu Y / Wu J | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2017 Journal: Mol Cell / Year: 2017Title: Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle. Authors: Ying Lu / Jiayi Wu / Yuanchen Dong / Shuobing Chen / Shuangwu Sun / Yong-Bei Ma / Qi Ouyang / Daniel Finley / Marc W Kirschner / Youdong Mao /   Abstract: The proteasome holoenzyme is activated by its regulatory particle (RP) consisting of two subcomplexes, the lid and the base. A key event in base assembly is the formation of a heterohexameric ring of ...The proteasome holoenzyme is activated by its regulatory particle (RP) consisting of two subcomplexes, the lid and the base. A key event in base assembly is the formation of a heterohexameric ring of AAA-ATPases, which is guided by at least four RP assembly chaperones in mammals: PAAF1, p28/gankyrin, p27/PSMD9, and S5b. Using cryogenic electron microscopy, we analyzed the non-AAA structure of the p28-bound human RP at 4.5 Å resolution and determined seven distinct conformations of the Rpn1-p28-AAA subcomplex within the p28-bound RP at subnanometer resolutions. Remarkably, the p28-bound AAA ring does not form a channel in the free RP and spontaneously samples multiple "open" and "closed" topologies at the Rpt2-Rpt6 and Rpt3-Rpt4 interfaces. Our analysis suggests that p28 assists the proteolytic core particle to select a specific conformation of the ATPase ring for RP engagement and is released in a shoehorn-like fashion in the last step of the chaperone-mediated proteasome assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8683.map.gz emd_8683.map.gz | 81.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8683-v30.xml emd-8683-v30.xml emd-8683.xml emd-8683.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8683.png emd_8683.png | 66.8 KB | ||
| Filedesc metadata |  emd-8683.cif.gz emd-8683.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8683 http://ftp.pdbj.org/pub/emdb/structures/EMD-8683 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8683 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8683 | HTTPS FTP |
-Related structure data
| Related structure data |  5vhrMC  8672C  8674C  8675C  8676C  8677C  8678C  8679C  8680C  8681C  8682C  8684C  5vgzC  5vhfC  5vhhC  5vhiC  5vhjC  5vhmC  5vhnC  5vhoC  5vhpC  5vhqC  5vhsC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10091 (Title: Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle EMPIAR-10091 (Title: Conformational Landscape of the p28-Bound Human Proteasome Regulatory ParticleData size: 70.0 Data #1: Classified single-particle datasets for multiple conformations of p28-bound human regulatory complex [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8683.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8683.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final Map of Rpn1-p28-AAA subcomplex in the TA7 state, corrected with a B-factor of -400 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
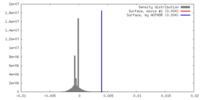
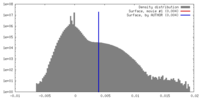
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Proteasome regulatory particle
| Entire | Name: Proteasome regulatory particle |
|---|---|
| Components |
|
-Supramolecule #1: Proteasome regulatory particle
| Supramolecule | Name: Proteasome regulatory particle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 26S proteasome non-ATPase regulatory subunit 10
| Macromolecule | Name: 26S proteasome non-ATPase regulatory subunit 10 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.14249 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CVSNLMVCNL AYSGKLEELK ESILADKSLA TRTDQDSRTA LHWACSAGHT EIVEFLLQLG VPVNDKDDAG WSPLHIAASA GRDEIVKAL LGKGAQVNAV NQNGCTPLHY AASKNRHEIA VMLLEGGANP DAKDHYEATA MHRAAAKGNL KMIHILLYYK A STNIQDTE ...String: CVSNLMVCNL AYSGKLEELK ESILADKSLA TRTDQDSRTA LHWACSAGHT EIVEFLLQLG VPVNDKDDAG WSPLHIAASA GRDEIVKAL LGKGAQVNAV NQNGCTPLHY AASKNRHEIA VMLLEGGANP DAKDHYEATA MHRAAAKGNL KMIHILLYYK A STNIQDTE GNTPLHLACD EERVEEAKLL VSQGASIYIE NKEEKTPLQV AKGGLGLILK RMVEG UniProtKB: 26S proteasome non-ATPase regulatory subunit 10 |
-Macromolecule #2: 26S proteasome regulatory subunit 7
| Macromolecule | Name: 26S proteasome regulatory subunit 7 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.746465 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PTVTMMQVEE KPDVTYSDVG GCKEQIEKLR EVVETPLLHP ERFVNLGIEP PKGVLLFGPP GTGKTLCARA VANRTDACFI RVIGSELVQ KYVGEGARMV RELFEMARTK KACLIFFDEI DAIGGARFDD GAGGDNEVQR TMLELINQLD GFDPRGNIKV L MATNRPDT ...String: PTVTMMQVEE KPDVTYSDVG GCKEQIEKLR EVVETPLLHP ERFVNLGIEP PKGVLLFGPP GTGKTLCARA VANRTDACFI RVIGSELVQ KYVGEGARMV RELFEMARTK KACLIFFDEI DAIGGARFDD GAGGDNEVQR TMLELINQLD GFDPRGNIKV L MATNRPDT LDPALMRPGR LDRKIEFSLP DLEGRTHIFK IHARSMSVER DIRFELLARL CPNSTGAEIR SVCTEAGMFA IR ARRKIAT EKDFLEAVNK VIKSYAKFS UniProtKB: 26S proteasome regulatory subunit 7 |
-Macromolecule #3: 26S proteasome regulatory subunit 4
| Macromolecule | Name: 26S proteasome regulatory subunit 4 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.958395 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TDPLVTVMKV EKAPQETYAD IGGLDNQIQE IKESVELPLT HPEYYEEMGI KPPKGVILYG PPGTGKTLLA KAVANQTSAT FLRVVGSEL IQKYLGDGPK LVRELFRVAE EHAPSIVFID EIDAIGTKRY DSNSGGEREI QRTMLELLNQ LDGFDSRGDV K VIMATNRI ...String: TDPLVTVMKV EKAPQETYAD IGGLDNQIQE IKESVELPLT HPEYYEEMGI KPPKGVILYG PPGTGKTLLA KAVANQTSAT FLRVVGSEL IQKYLGDGPK LVRELFRVAE EHAPSIVFID EIDAIGTKRY DSNSGGEREI QRTMLELLNQ LDGFDSRGDV K VIMATNRI ETLDPALIRP GRIDRKIEFP LPDEKTKKRI FQIHTSRMTL ADDVTLDDLI MAKDDLSGAD IKAICTEAGL MA LRERRMK VTNEDFKKSK ENVLYKKQEG UniProtKB: 26S proteasome regulatory subunit 4 |
-Macromolecule #4: 26S proteasome regulatory subunit 6B
| Macromolecule | Name: 26S proteasome regulatory subunit 6B / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.497975 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PEADSSIMML TSDQKPDVMY ADIGGMDIQK QEVREAVELP LTHFELYKQI GIDPPRGVLM YGPPGCGKTM LAKAVAHHTT AAFIRVVGS EFVQKYLGEG PRMVRDVFRL AKENAPAIIF IDEIDAIATK RFDAQTGADR EVQRILLELL NQMDGFDQNV N VKVIMATN ...String: PEADSSIMML TSDQKPDVMY ADIGGMDIQK QEVREAVELP LTHFELYKQI GIDPPRGVLM YGPPGCGKTM LAKAVAHHTT AAFIRVVGS EFVQKYLGEG PRMVRDVFRL AKENAPAIIF IDEIDAIATK RFDAQTGADR EVQRILLELL NQMDGFDQNV N VKVIMATN RADTLDPALL RPGRLDRKIE FPLPDRRQKR LIFSTITSKM NLSEEVDLED YVARPDKISG ADINSICQES GM LAVRENR YIVLAKDFEK AYKTV UniProtKB: 26S proteasome regulatory subunit 6B |
-Macromolecule #5: 26S proteasome regulatory subunit 10B
| Macromolecule | Name: 26S proteasome regulatory subunit 10B / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.434889 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GNVSYSEIGG LSEQIRELRE VIELPLTNPE LFQRVGIIPP KGCLLYGPPG TGKTLLARAV ASQLDCNFLK VVSSSIVDKY IGESARLIR EMFNYARDHQ PCIIFMDEID AIGGRRFSEG TSADREIQRT LMELLNQMDG FDTLHRVKMI MATNRPDTLD P ALLRPGRL ...String: GNVSYSEIGG LSEQIRELRE VIELPLTNPE LFQRVGIIPP KGCLLYGPPG TGKTLLARAV ASQLDCNFLK VVSSSIVDKY IGESARLIR EMFNYARDHQ PCIIFMDEID AIGGRRFSEG TSADREIQRT LMELLNQMDG FDTLHRVKMI MATNRPDTLD P ALLRPGRL DRKIHIDLPN EQARLDILKI HAGPITKHGE IDYEAIVKLS DGFNGADLRN VCTEAGMFAI RADHDFVVQE DF MKAVRKV ADSKKLESKL DYKPV UniProtKB: 26S proteasome regulatory subunit 10B |
-Macromolecule #6: 26S proteasome regulatory subunit 6A
| Macromolecule | Name: 26S proteasome regulatory subunit 6A / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.773246 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TEYDSRVKAM EVDERPTEQY SDIGGLDKQI QELVEAIVLP MNHKEKFENL GIQPPKGVLM YGPPGTGKTL LARACAAQTK ATFLKLAGP QLVQMFIGDG AKLVRDAFAL AKEKAPSIIF IDELDAIGTK RFDSEKAGDR EVQRTMLELL NQLDGFQPNT Q VKVIAATN ...String: TEYDSRVKAM EVDERPTEQY SDIGGLDKQI QELVEAIVLP MNHKEKFENL GIQPPKGVLM YGPPGTGKTL LARACAAQTK ATFLKLAGP QLVQMFIGDG AKLVRDAFAL AKEKAPSIIF IDELDAIGTK RFDSEKAGDR EVQRTMLELL NQLDGFQPNT Q VKVIAATN RVDILDPALL RSGRLDRKIE FPMPNEEARA RIMQIHSRKM NVSPDVNYEE LARCTDDFNG AQCKAVCVEA GM IALRRGA TELTHEDYME GILEVQAKKK UniProtKB: 26S proteasome regulatory subunit 6A |
-Macromolecule #7: 26S proteasome regulatory subunit 8
| Macromolecule | Name: 26S proteasome regulatory subunit 8 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.501363 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KVDPLVSLMM VEKVPDSTYE MIGGLDKQIK EIKEVIELPV KHPELFEALG IAQPKGVLLY GPPGTGKTLL ARAVAHHTDC TFIRVSGSE LVQKFIGEGA RMVRELFVMA REHAPSIIFM DEIDSIGSSR LEGGSGGDSE VQRTMLELLN QLDGFEATKN I KVIMATNR ...String: KVDPLVSLMM VEKVPDSTYE MIGGLDKQIK EIKEVIELPV KHPELFEALG IAQPKGVLLY GPPGTGKTLL ARAVAHHTDC TFIRVSGSE LVQKFIGEGA RMVRELFVMA REHAPSIIFM DEIDSIGSSR LEGGSGGDSE VQRTMLELLN QLDGFEATKN I KVIMATNR IDILDSALLR PGRIDRKIEF PPPNEEARLD ILKIHSRKMN LTRGINLRKI AELMPGASGA EVKGVCTEAG MY ALRERRV HVTQEDFEMA VAKVMQKDS UniProtKB: 26S proteasome regulatory subunit 8 |
-Macromolecule #8: 26S proteasome non-ATPase regulatory subunit 2
| Macromolecule | Name: 26S proteasome non-ATPase regulatory subunit 2 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.790188 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RDKAPVQPQQ SPAAAPGGTD EKPSGKERRD AGDKDKEQEL SEEDKQLQDE LEMLVERLGE KDTSLYRPAL EELRRQIRSS TTSMTSVPK PLKFLRPHYG KLKEIYENMA PGENKRFAAD IISVLAMTMS GERECLKYRL VGSQEELASW GHEYVRHLAG E VAKEWQEL ...String: RDKAPVQPQQ SPAAAPGGTD EKPSGKERRD AGDKDKEQEL SEEDKQLQDE LEMLVERLGE KDTSLYRPAL EELRRQIRSS TTSMTSVPK PLKFLRPHYG KLKEIYENMA PGENKRFAAD IISVLAMTMS GERECLKYRL VGSQEELASW GHEYVRHLAG E VAKEWQEL DDAEKVQREP LLTLVKEIVP YNMAHNAEHE ACDLLMEIEQ VDMLEKDIDE NAYAKVCLYL TSCVNYVPEP EN SALLRCA LGVFRKFSRF PEALRLALML NDMELVEDIF TSCKDVVVQK QMAFMLGRHG VFLELSEDVE EYEDLTEIMS NVQ LNSNFL ALARELDIME PKVPDDIYKT HLENNRFGGS GSQVDSARMN LASSFVNGFV NAAFGQDKLL TDDGNKWLYK NKDH GMLSA AASLGMILLW DVDGGLTQID KYLYSSEDYI KSGALLACGI VNSGVRNECD PALALLSDYV LHNSNTMRLG SIFGL GLAY AGSNREDVLT LLLPVMGDSK SSMEVAGVTA LACGMIAVGS CNGDVTSTIL QTIMEKSETE LKDTYARWLP LGLGLN HLG KGEAIEAILA ALEVVSEPFR SFANTLVDVC AYAGSGNVLK VQQLLHICSE HFDSKEKEED KDKKEKKDKD KKEAPAD MG AHQGVAVLGI ALIAMGEEIG AEMALRTFGH LLRYGEPTLR RAVPLALALI SVSNPRLNIL DTLSKFSHDA DPEVSYNS I FAMGMVGSGT NNARLAAMLR QLAQYHAKDP NNLFMVRLAQ GLTHLGKGTL TLCPYHSDRQ LMSQVAVAGL LTVLVSFLD VRNIILGKSH YVLYGLVAAM QPRMLVTFDE ELRPLPVSVR VGQAVDVV UniProtKB: 26S proteasome non-ATPase regulatory subunit 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 14464 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)