+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8477 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRISPR RNA-guided surveillance complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / Cascade / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / defense response to virus / RNA binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   Thermobifida fusca (strain YX) (bacteria) / Thermobifida fusca (strain YX) (bacteria) /   Thermobifida fusca YX (bacteria) Thermobifida fusca YX (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Xiao Y / Luo M | |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System. Authors: Yibei Xiao / Min Luo / Robert P Hayes / Jonathan Kim / Sherwin Ng / Fang Ding / Maofu Liao / Ailong Ke /  Abstract: Type I CRISPR systems feature a sequential dsDNA target searching and degradation process, by crRNA-displaying Cascade and nuclease-helicase fusion enzyme Cas3, respectively. Here we present two cryo- ...Type I CRISPR systems feature a sequential dsDNA target searching and degradation process, by crRNA-displaying Cascade and nuclease-helicase fusion enzyme Cas3, respectively. Here we present two cryo-EM snapshots of the Thermobifida fusca type I-E Cascade: (1) unwinding 11 bp of dsDNA at the seed-sequence region to scout for sequence complementarity, and (2) further unwinding of the entire protospacer to form a full R-loop. These structures provide the much-needed temporal and spatial resolution to resolve key mechanistic steps leading to Cas3 recruitment. In the early steps, PAM recognition causes severe DNA bending, leading to spontaneous DNA unwinding to form a seed-bubble. The full R-loop formation triggers conformational changes in Cascade, licensing Cas3 to bind. The same process also generates a bulge in the non-target DNA strand, enabling its handover to Cas3 for cleavage. The combination of both negative and positive checkpoints ensures stringent yet efficient target degradation in type I CRISPR-Cas systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8477.map.gz emd_8477.map.gz | 58.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8477-v30.xml emd-8477-v30.xml emd-8477.xml emd-8477.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8477.png emd_8477.png | 54.8 KB | ||
| Filedesc metadata |  emd-8477.cif.gz emd-8477.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8477 http://ftp.pdbj.org/pub/emdb/structures/EMD-8477 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8477 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8477 | HTTPS FTP |
-Related structure data
| Related structure data |  5u07MC  8478C  5u0aC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8477.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8477.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
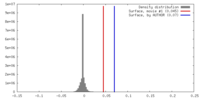
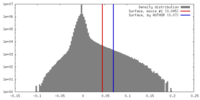
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CRISPR RNA-guided surveillance complex
| Entire | Name: CRISPR RNA-guided surveillance complex |
|---|---|
| Components |
|
-Supramolecule #1: CRISPR RNA-guided surveillance complex
| Supramolecule | Name: CRISPR RNA-guided surveillance complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca (strain YX) (bacteria) Thermobifida fusca (strain YX) (bacteria) |
-Macromolecule #1: CRISPR-associated protein, Cse3 family
| Macromolecule | Name: CRISPR-associated protein, Cse3 family / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) / Strain: YX Thermobifida fusca YX (bacteria) / Strain: YX |
| Molecular weight | Theoretical: 26.327938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTWLTKIVPD LRYRQTRADF RTAGNLHRKL IRLSSDLGEE RIANPRQQSG LLFRIEETRN ELYLLVQSHS PLRVDRLGPG YHGVQMRNL DPFLARLDKG SRVRYRIVAS PTKRLGRSEN NTQRLGLKEP PKKPREYTWA LRGAAAEEWW HSRAAANGLE L LSTYAQTL ...String: MTWLTKIVPD LRYRQTRADF RTAGNLHRKL IRLSSDLGEE RIANPRQQSG LLFRIEETRN ELYLLVQSHS PLRVDRLGPG YHGVQMRNL DPFLARLDKG SRVRYRIVAS PTKRLGRSEN NTQRLGLKEP PKKPREYTWA LRGAAAEEWW HSRAAANGLE L LSTYAQTL DDVRDPGTAD RSRKIRHPAV RFDGEAVISD VDAVRHAVLN GIGRGKSYGC GLLSLALIEE GEHG UniProtKB: CRISPR-associated protein, Cse3 family |
-Macromolecule #2: Cse2
| Macromolecule | Name: Cse2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) / Strain: YX Thermobifida fusca YX (bacteria) / Strain: YX |
| Molecular weight | Theoretical: 27.446613 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNSDYILQHA DALVKRVSKL IVNEPAARAA LRRGVGLAPE DPRMLAAHRV VAPYVPVPTD YDVDRRRAAS LWDVHAVERA FYAVAAIMA AQPRSARDQE AEATEEQTGE PQDSEALTEP TPAEESSATK DGKPDRRPNL GVSLAQAVFD KGLNADSTEQ R LHLIARQN ...String: MNSDYILQHA DALVKRVSKL IVNEPAARAA LRRGVGLAPE DPRMLAAHRV VAPYVPVPTD YDVDRRRAAS LWDVHAVERA FYAVAAIMA AQPRSARDQE AEATEEQTGE PQDSEALTEP TPAEESSATK DGKPDRRPNL GVSLAQAVFD KGLNADSTEQ R LHLIARQN LDGVHRHLPR LVLYLRSDQV HIDWGILIRD LARWGHTPRH VAREWVQDYH RTLETLTRQA EQKNKNNTTD EE AEAA UniProtKB: CRISPR-associated protein, Cse2 family |
-Macromolecule #3: CRISPR-associated protein, Cse1 family
| Macromolecule | Name: CRISPR-associated protein, Cse1 family / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) / Strain: YX Thermobifida fusca YX (bacteria) / Strain: YX |
| Molecular weight | Theoretical: 61.433297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLSVALCFLV GGAIPSPPSF DVTIAPWLIA RSRDVLAAPE MLGLRDVLIR SHELSDVEIP LPPGAAVLWR ILALITARIT GLDQPPNKN PKRKWQARRS QILSKGRLDP EAVDAYFADY SERFDLFHPE RPWLQDPRLR EECPKTSGVN KLAWGRTAGE N QVWLGGHH ...String: MLSVALCFLV GGAIPSPPSF DVTIAPWLIA RSRDVLAAPE MLGLRDVLIR SHELSDVEIP LPPGAAVLWR ILALITARIT GLDQPPNKN PKRKWQARRS QILSKGRLDP EAVDAYFADY SERFDLFHPE RPWLQDPRLR EECPKTSGVN KLAWGRTAGE N QVWLGGHH HDLDPHPLDS AEAVWHLLAT LGYGPSGMCT ARVVRGRSER NVTAGPLRGT VSYHPLGRTL FESLILNIPY PG TGAADLA FWEQPELNDP LGLPEESAGL AGILRLDHFR HAVLLHPSPD GSHVVDAWVT WAWRERNISP ELDPYLIYQT SKE GRVYPR PAEAERAIWR DLDALLHYGE DGNYRPTILD NCTPLAQVPQ EVLDSLRLRA FGFDQDGQAR DKQWFTATTP AVLR WLADR ETDDNENARI VRRITLARKA AEALGRRLEK ACKEAWKESN SPSSTSSGTN AKTETGVGPW VQHGMSRYWA KAEPV FWNI VYDRPAQGYT PGMAGPGNAF NLVALAAYDE VTGPYCERPR VAKVVERHRS TLFSNWTPKQ DKEAA UniProtKB: CRISPR-associated protein, Cse1 family |
-Macromolecule #4: CRISPR-associated protein, Cse4 family
| Macromolecule | Name: CRISPR-associated protein, Cse4 family / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) / Strain: YX Thermobifida fusca YX (bacteria) / Strain: YX |
| Molecular weight | Theoretical: 41.043043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTFVDIHAIQ TLPYSNINRD DLGSPKTVVY GGKERTRVSS QSWKRAVRHE VEARLGDKAV RTRRIISEIA KRLRERGWDA DLADAGARQ VVLSVGKKSG IKLEKEKDSE APATSVLFYL PVPAIDELAA IADEHRDAVA KEAAKKTPKG ILPADRITEV L KSRNVSVN ...String: MTFVDIHAIQ TLPYSNINRD DLGSPKTVVY GGKERTRVSS QSWKRAVRHE VEARLGDKAV RTRRIISEIA KRLRERGWDA DLADAGARQ VVLSVGKKSG IKLEKEKDSE APATSVLFYL PVPAIDELAA IADEHRDAVA KEAAKKTPKG ILPADRITEV L KSRNVSVN LFGRMLAELP STEVDGAVQF AHAFTVHGTT VEVDFFTAVD DIPKENDHGS GHMNAGQFSA GTFYRYANVN LD RLVENTG DAQTARTAVA EFLRAFLSTV PSGKQNATAA MTLPDLVHIA VRFDRPISFA PAFETALYGS DGYTLRACQE LNN YAERLR EVWPDDAIRG YATVENKTDL AALGERYDSY PALIDAMVAA AFEGERE UniProtKB: CRISPR-associated protein, Cse4 family |
-Macromolecule #7: CRISPR-associated protein, Cas5e family
| Macromolecule | Name: CRISPR-associated protein, Cas5e family / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) / Strain: YX Thermobifida fusca YX (bacteria) / Strain: YX |
| Molecular weight | Theoretical: 28.27926 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGFLLRLAG PMQSWGEHSM FGERDTLPYP SRSGLIGMFA AAQGVRRGDP LDRYKELKFT VRVDRPGVRL VDFHTIGGGL PKERTVPTA AGERRDPKKA TIVTSRSYLA DAVFTVAVTG PEADTIADAL AAPYWQPYLG RRAFVPDPLL VLRRRVADPV R ELVEAVPL ...String: MSGFLLRLAG PMQSWGEHSM FGERDTLPYP SRSGLIGMFA AAQGVRRGDP LDRYKELKFT VRVDRPGVRL VDFHTIGGGL PKERTVPTA AGERRDPKKA TIVTSRSYLA DAVFTVAVTG PEADTIADAL AAPYWQPYLG RRAFVPDPLL VLRRRVADPV R ELVEAVPL PHRRVEEDAA TVLVDLIYEE GEYPDTRTLT VLNDVPLSFD SKSRRYSTRQ IRVVPTEVPA TLVAGPGRDY QN KLFTYVK QCAEEAA UniProtKB: CRISPR-associated protein, Cas5e family |
-Macromolecule #5: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) Thermobifida fusca YX (bacteria) |
| Molecular weight | Theoretical: 19.790793 KDa |
| Sequence | String: AUGGACCGCC AGUGAUAAGU GGAAUGCCAU GUGGGCUGUC GUGAGCCCCA CGCACGUGGG G |
-Macromolecule #6: Target Strand
| Macromolecule | Name: Target Strand / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) Thermobifida fusca YX (bacteria) |
| Molecular weight | Theoretical: 6.381106 KDa |
| Sequence | String: (DT)(DT)(DA)(DT)(DC)(DA)(DC)(DT)(DG)(DG) (DC)(DT)(DT)(DC)(DG)(DT)(DC)(DC)(DG)(DC) (DG) |
-Macromolecule #8: Nontarget Strand
| Macromolecule | Name: Nontarget Strand / type: dna / ID: 8 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Thermobifida fusca YX (bacteria) Thermobifida fusca YX (bacteria) |
| Molecular weight | Theoretical: 3.9866 KDa |
| Sequence | String: (DC)(DG)(DC)(DG)(DG)(DA)(DC)(DG)(DA)(DA) (DG)(DC)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 10 mM Hepes, 150 mM NaCl, 5 mM DTT, pH 7.5 |
| Grid | Model: 400 mesh Quantifoil holey carbon grid / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80.0 K / Max: 105.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1305 / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.3000000000000003 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 31000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)