[English] 日本語
 Yorodumi
Yorodumi- EMDB-8472: MicroED structure of a complex between monomeric TGF-b and its re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8472 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | |||||||||
 Map data Map data | Complex between monomeric TGF-b and its receptor, TbRII | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of timing of catagen / regulation of apoptotic process involved in outflow tract morphogenesis / negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation / substantia propria of cornea development / ascending aorta morphogenesis / positive regulation of activation-induced cell death of T cells / cardioblast differentiation / positive regulation of tolerance induction to self antigen / positive regulation of B cell tolerance induction / uterine wall breakdown ...regulation of timing of catagen / regulation of apoptotic process involved in outflow tract morphogenesis / negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation / substantia propria of cornea development / ascending aorta morphogenesis / positive regulation of activation-induced cell death of T cells / cardioblast differentiation / positive regulation of tolerance induction to self antigen / positive regulation of B cell tolerance induction / uterine wall breakdown / inferior endocardial cushion morphogenesis / transforming growth factor beta receptor activity, type II / bronchus morphogenesis / positive regulation of timing of catagen / mammary gland morphogenesis / lens fiber cell apoptotic process / growth plate cartilage chondrocyte growth / positive regulation of cardioblast differentiation / tricuspid valve morphogenesis / TGFBR2 MSI Frameshift Mutants in Cancer / cardiac right ventricle morphogenesis / miRNA transport / regulation of transforming growth factor beta2 production / transforming growth factor beta ligand-receptor complex / atrial septum morphogenesis / pharyngeal arch artery morphogenesis / positive regulation of heart contraction / aorta morphogenesis / type III transforming growth factor beta receptor binding / positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation / Langerhans cell differentiation / TGFBR2 Kinase Domain Mutants in Cancer / transforming growth factor beta receptor activity / activation-induced cell death of T cells / glial cell migration / cardiac left ventricle morphogenesis / positive regulation of extracellular matrix disassembly / secondary palate development / negative regulation of macrophage cytokine production / SMAD2/3 Phosphorylation Motif Mutants in Cancer / TGFBR1 KD Mutants in Cancer / somatic stem cell division / positive regulation of integrin biosynthetic process / atrial septum primum morphogenesis / endocardial cushion fusion / membranous septum morphogenesis / positive regulation of T cell tolerance induction / heart valve morphogenesis / TGFBR3 regulates TGF-beta signaling / positive regulation of NK T cell differentiation / negative regulation of cartilage development / cardiac epithelial to mesenchymal transition / signaling / positive regulation of stress-activated MAPK cascade / neuron fate commitment / pericyte cell differentiation / activin receptor complex / activin receptor activity, type I / embryonic digestive tract development / lung lobe morphogenesis / transforming growth factor beta receptor binding / eye development / neural retina development / type II transforming growth factor beta receptor binding / transmembrane receptor protein serine/threonine kinase activity / receptor protein serine/threonine kinase / regulation of stem cell proliferation / pulmonary valve morphogenesis / activin binding / cranial skeletal system development / TGFBR1 LBD Mutants in Cancer / SMAD protein signal transduction / type I transforming growth factor beta receptor binding / myeloid dendritic cell differentiation / embryonic cranial skeleton morphogenesis / activin receptor signaling pathway / glycosaminoglycan binding / ventricular trabecula myocardium morphogenesis / negative regulation of Ras protein signal transduction / positive regulation of CD4-positive, alpha-beta T cell proliferation / embryo development ending in birth or egg hatching / regulation of stem cell differentiation / response to cholesterol / outflow tract septum morphogenesis / positive regulation of extrinsic apoptotic signaling pathway in absence of ligand / cell-cell junction organization / transforming growth factor beta binding / collagen fibril organization / kinase activator activity / embryonic limb morphogenesis / aortic valve morphogenesis / lens development in camera-type eye / positive regulation of cell adhesion mediated by integrin / atrioventricular valve morphogenesis / face morphogenesis / odontogenesis / embryonic hemopoiesis / positive regulation of mesenchymal cell proliferation / artery morphogenesis / endocardial cushion morphogenesis Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron crystallography / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Weiss SC / de la Cruz MJ | |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2017 Journal: Nat Methods / Year: 2017Title: Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Authors: M Jason de la Cruz / Johan Hattne / Dan Shi / Paul Seidler / Jose Rodriguez / Francis E Reyes / Michael R Sawaya / Duilio Cascio / Simon C Weiss / Sun Kyung Kim / Cynthia S Hinck / Andrew P ...Authors: M Jason de la Cruz / Johan Hattne / Dan Shi / Paul Seidler / Jose Rodriguez / Francis E Reyes / Michael R Sawaya / Duilio Cascio / Simon C Weiss / Sun Kyung Kim / Cynthia S Hinck / Andrew P Hinck / Guillermo Calero / David Eisenberg / Tamir Gonen /  Abstract: Traditionally, crystallographic analysis of macromolecules has depended on large, well-ordered crystals, which often require significant effort to obtain. Even sizable crystals sometimes suffer from ...Traditionally, crystallographic analysis of macromolecules has depended on large, well-ordered crystals, which often require significant effort to obtain. Even sizable crystals sometimes suffer from pathologies that render them inappropriate for high-resolution structure determination. Here we show that fragmentation of large, imperfect crystals into microcrystals or nanocrystals can provide a simple path for high-resolution structure determination by the cryoEM method MicroED and potentially by serial femtosecond crystallography. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8472.map.gz emd_8472.map.gz | 604.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8472-v30.xml emd-8472-v30.xml emd-8472.xml emd-8472.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8472.png emd_8472.png | 372.9 KB | ||
| Filedesc metadata |  emd-8472.cif.gz emd-8472.cif.gz | 5.4 KB | ||
| Filedesc structureFactors |  emd_8472_sf.cif.gz emd_8472_sf.cif.gz | 955.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8472 http://ftp.pdbj.org/pub/emdb/structures/EMD-8472 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8472 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8472 | HTTPS FTP |
-Validation report
| Summary document |  emd_8472_validation.pdf.gz emd_8472_validation.pdf.gz | 476.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8472_full_validation.pdf.gz emd_8472_full_validation.pdf.gz | 475.7 KB | Display | |
| Data in XML |  emd_8472_validation.xml.gz emd_8472_validation.xml.gz | 4.1 KB | Display | |
| Data in CIF |  emd_8472_validation.cif.gz emd_8472_validation.cif.gz | 4.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8472 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8472 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8472 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8472 | HTTPS FTP |
-Related structure data
| Related structure data |  5ty4MC  8216C  8217C  8218C  8219C  8220C  8221C  8222C  5k7nC  5k7oC  5k7pC  5k7qC  5k7rC  5k7sC  5k7tC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8472.map.gz / Format: CCP4 / Size: 846.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8472.map.gz / Format: CCP4 / Size: 846.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex between monomeric TGF-b and its receptor, TbRII | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.94386 Å / Y: 0.93855 Å / Z: 0.94653 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
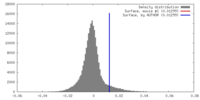
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 19 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex between monomeric TGF-b and its receptor, TbRII
| Entire | Name: Complex between monomeric TGF-b and its receptor, TbRII |
|---|---|
| Components |
|
-Supramolecule #1: Complex between monomeric TGF-b and its receptor, TbRII
| Supramolecule | Name: Complex between monomeric TGF-b and its receptor, TbRII type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 19.072 KDa |
-Macromolecule #1: TGF-beta receptor type-2
| Macromolecule | Name: TGF-beta receptor type-2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: receptor protein serine/threonine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.788519 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FPQLCKFCDV RFSTCDNQKS CMSNCSITSI CEKPQEVCVA VWRKNDENIT LETVCHDPKL PYHDFILEDA ASPTCIMKEK KKPGETFFM CSCSSDECND NIIF UniProtKB: TGF-beta receptor type-2 |
-Macromolecule #2: mmTGF-b2-7m
| Macromolecule | Name: mmTGF-b2-7m / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.076813 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CCLRPLYIDF RKDLGWKWIH EPKGYNANFC AGACPYLWSS DTQHSRVLSL YNTINPEASA SPCCVSQDLE PLTIVYYVGR KPKVEQLSN MIVKSCKC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component - Concentration: 100.0 mM / Component - Name: HEPES |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number grids imaged: 2 / Number real images: 353 / Number diffraction images: 353 / Average exposure time: 4.1 sec. / Average electron dose: 0.004 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Camera length: 2000 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-5ty4: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)