[English] 日本語
 Yorodumi
Yorodumi- EMDB-8419: High affinity anchoring of the decoration protein pb10 onto the b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8419 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid | |||||||||
 Map data Map data | decorated empty expanded capsid of bacteriophage T5 | |||||||||
 Sample Sample | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid != Escherichia phage T5 High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid
| |||||||||
| Function / homology | viral scaffold / T=13 icosahedral viral capsid / : / Phage capsid / Phage capsid family / symbiont-mediated evasion of host immune response / viral capsid / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Escherichia phage T5 (virus) Escherichia phage T5 (virus) | |||||||||
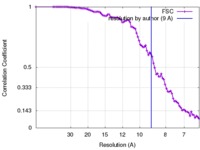
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Conway J / Huet A | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2017 Journal: Sci Rep / Year: 2017Title: High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid. Authors: Emeline Vernhes / Madalena Renouard / Bernard Gilquin / Philippe Cuniasse / Dominique Durand / Patrick England / Sylviane Hoos / Alexis Huet / James F Conway / Anatoly Glukhov / Vladimir ...Authors: Emeline Vernhes / Madalena Renouard / Bernard Gilquin / Philippe Cuniasse / Dominique Durand / Patrick England / Sylviane Hoos / Alexis Huet / James F Conway / Anatoly Glukhov / Vladimir Ksenzenko / Eric Jacquet / Naïma Nhiri / Sophie Zinn-Justin / Pascale Boulanger /    Abstract: Bacteriophage capsids constitute icosahedral shells of exceptional stability that protect the viral genome. Many capsids display on their surface decoration proteins whose structure and function ...Bacteriophage capsids constitute icosahedral shells of exceptional stability that protect the viral genome. Many capsids display on their surface decoration proteins whose structure and function remain largely unknown. The decoration protein pb10 of phage T5 binds at the centre of the 120 hexamers formed by the major capsid protein. Here we determined the 3D structure of pb10 and investigated its capsid-binding properties using NMR, SAXS, cryoEM and SPR. Pb10 consists of an α-helical capsid-binding domain and an Ig-like domain exposed to the solvent. It binds to the T5 capsid with a remarkably high affinity and its binding kinetics is characterized by a very slow dissociation rate. We propose that the conformational exchange events observed in the capsid-binding domain enable rearrangements upon binding that contribute to the quasi-irreversibility of the pb10-capsid interaction. Moreover we show that pb10 binding is a highly cooperative process, which favours immediate rebinding of newly dissociated pb10 to the 120 hexamers of the capsid protein. In extreme conditions, pb10 protects the phage from releasing its genome. We conclude that pb10 may function to reinforce the capsid thus favouring phage survival in harsh environments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8419.map.gz emd_8419.map.gz | 161.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8419-v30.xml emd-8419-v30.xml emd-8419.xml emd-8419.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8419_fsc.xml emd_8419_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_8419.png emd_8419.png | 269.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8419 http://ftp.pdbj.org/pub/emdb/structures/EMD-8419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8419 | HTTPS FTP |
-Related structure data
| Related structure data |  5tjtMC  8423C  5lxkC  5lxlC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8419.map.gz / Format: CCP4 / Size: 479.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8419.map.gz / Format: CCP4 / Size: 479.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | decorated empty expanded capsid of bacteriophage T5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.74 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
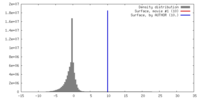
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : High affinity anchoring of the decoration protein pb10 onto the b...
| Entire | Name: High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage T5
| Supramolecule | Name: Escherichia phage T5 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 10726 / Sci species name: Escherichia phage T5 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism: |
| Virus shell | Shell ID: 1 / Name: expanded head / Diameter: 900.0 Å / T number (triangulation number): 13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||
| Grid | Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 1939 / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 6.16 µm / Nominal defocus min: 1.27 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5tjt: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)