+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8325 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
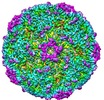
| Title | Beta-propiolactone treated coxsackievirus A16 empty procapsid | |||||||||
 Map data Map data | Beta-propiolactone treated coxsackievirus A16 empty procapsid | |||||||||
 Sample Sample | coxsackievirus A16 capsids != COXSACKIEVIRUS A16 coxsackievirus A16 capsids
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |   COXSACKIEVIRUS A16 COXSACKIEVIRUS A16 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | Chen F / Yao C | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2017 Journal: J Virol / Year: 2017Title: Beta-Propiolactone Inactivation of Coxsackievirus A16 Induces Structural Alteration and Surface Modification of Viral Capsids. Authors: Chen Fan / Xiaohua Ye / Zhiqiang Ku / Liangliang Kong / Qingwei Liu / Cong Xu / Yao Cong / Zhong Huang /  Abstract: Beta-propiolactone (BPL) is an inactivating agent that is widely used in the vaccine industry. However, its effects on vaccine protein antigens and its mechanisms of action remain poorly understood. ...Beta-propiolactone (BPL) is an inactivating agent that is widely used in the vaccine industry. However, its effects on vaccine protein antigens and its mechanisms of action remain poorly understood. Here we present cryo-electron microscopy (cryo-EM) structures of BPL-treated coxsackievirus A16 (CVA16) mature virions and procapsids at resolutions of 3.9 Å and 6.5 Å, respectively. Notably, both particles were found to adopt an expanded conformation resembling the 135S-like uncoating intermediate, with characteristic features including an opened 2-fold channel, the externalization of the N terminus of VP1 capsid protein, and the absence of pocket factor. However, major neutralizing epitopes are very well preserved on these particles. Further biochemical analyses revealed that BPL treatment impairs the abilities of CVA16 particles to bind to the attachment receptor heparan sulfate and to a conformation-dependent monoclonal antibody in a BPL dose-dependent manner, indicating that BPL is able to modify surface-exposed amino acid residues. Taken together, our results demonstrate that BPL treatment may induce alteration of the overall structure and surface properties of a nonenveloped viral capsid, thus revealing a novel mode of action of BPL. Beta-propiolactone (BPL) is commonly used as an inactivating reagent to produce viral vaccines. It is recognized that BPL inactivates viral infectivity through modification of viral nucleic acids. However, its effect on viral proteins remains largely unknown. Here, we present high-resolution cryo-EM structures of BPL-treated coxsackievirus A16 (CVA16) mature virions and procapsids, which reveals an expanded overall conformation and characteristic features that are typical for the 135S-like uncoating intermediate. We further show that the BPL concentration affects the binding of inactivated CVA16 particles to their receptor/antibody. Thus, BPL treatment can alter the overall structure and surface properties of viral capsids, which may lead to antigenic and immunogenic variations. Our findings provide important information for future development of BPL-inactivated vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8325.map.gz emd_8325.map.gz | 192.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8325-v30.xml emd-8325-v30.xml emd-8325.xml emd-8325.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8325.png emd_8325.png | 313.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8325 http://ftp.pdbj.org/pub/emdb/structures/EMD-8325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8325 | HTTPS FTP |
-Related structure data
| Related structure data |  5tskMC  5tslMC  8324C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8325.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8325.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Beta-propiolactone treated coxsackievirus A16 empty procapsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : coxsackievirus A16 capsids
| Entire | Name: coxsackievirus A16 capsids |
|---|---|
| Components |
|
-Supramolecule #1: COXSACKIEVIRUS A16
| Supramolecule | Name: COXSACKIEVIRUS A16 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 31704 / Sci species name: COXSACKIEVIRUS A16 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.0032 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Component - Concentration: 0.15 M / Component - Formula: PBS / Component - Name: phosphate buffered saline |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK III / Details: blot for 2 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 90.0 K |
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Frames/image: 3-18 / Number grids imaged: 1 / Number real images: 1350 / Average exposure time: 1.1 sec. / Average electron dose: 25.0 e/Å2 Details: Images were recorded on a Falcon II direct electron detector in the 18-frame movie mode |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.007 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 57000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)