[English] 日本語
 Yorodumi
Yorodumi- EMDB-7850: Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7850 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | |||||||||
 Map data Map data | RAG in complex with 12-RSS and 23-RSS substrate DNAs, sharpened map with B factor -190 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | V(D)J recombination / RAG complex / unmelted DNA substrates / Pre-cleavage complex / Recombination-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsomatic diversification of immune receptors via germline recombination within a single locus / hematopoietic or lymphoid organ development / protein-DNA complex assembly / DNA recombinase complex / endodeoxyribonuclease complex / lymphocyte differentiation / immunoglobulin V(D)J recombination / V(D)J recombination / phosphatidylinositol-3,4-bisphosphate binding / histone H3K4me3 reader activity ...somatic diversification of immune receptors via germline recombination within a single locus / hematopoietic or lymphoid organ development / protein-DNA complex assembly / DNA recombinase complex / endodeoxyribonuclease complex / lymphocyte differentiation / immunoglobulin V(D)J recombination / V(D)J recombination / phosphatidylinositol-3,4-bisphosphate binding / histone H3K4me3 reader activity / phosphatidylinositol-3,5-bisphosphate binding / detection of maltose stimulus / maltose transport complex / phosphatidylinositol-3,4,5-trisphosphate binding / carbohydrate transport / T cell differentiation / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / phosphatidylinositol-4,5-bisphosphate binding / phosphatidylinositol binding / ATP-binding cassette (ABC) transporter complex / B cell differentiation / thymus development / cell chemotaxis / RING-type E3 ubiquitin transferase / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / T cell differentiation in thymus / outer membrane-bounded periplasmic space / chromatin organization / endonuclease activity / histone binding / DNA recombination / sequence-specific DNA binding / Hydrolases; Acting on ester bonds / adaptive immune response / periplasmic space / DNA damage response / chromatin binding / magnesium ion binding / protein homodimerization activity / DNA binding / zinc ion binding / metal ion binding / membrane / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Wu H / Liao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: DNA melting initiates the RAG catalytic pathway. Authors: Heng Ru / Wei Mi / Pengfei Zhang / Frederick W Alt / David G Schatz / Maofu Liao / Hao Wu /  Abstract: The mechanism for initiating DNA cleavage by DDE-family enzymes, including the RAG endonuclease, which initiates V(D)J recombination, is not well understood. Here we report six cryo-EM structures of ...The mechanism for initiating DNA cleavage by DDE-family enzymes, including the RAG endonuclease, which initiates V(D)J recombination, is not well understood. Here we report six cryo-EM structures of zebrafish RAG in complex with one or two intact recombination signal sequences (RSSs), at up to 3.9-Å resolution. Unexpectedly, these structures reveal DNA melting at the heptamer of the RSSs, thus resulting in a corkscrew-like rotation of coding-flank DNA and the positioning of the scissile phosphate in the active site. Substrate binding is associated with dimer opening and a piston-like movement in RAG1, first outward to accommodate unmelted DNA and then inward to wedge melted DNA. These precleavage complexes show limited base-specific contacts of RAG at the conserved terminal CAC/GTG sequence of the heptamer, thus suggesting conservation based on a propensity to unwind. CA and TG overwhelmingly dominate terminal sequences in transposons and retrotransposons, thereby implicating a universal mechanism for DNA melting during the initiation of retroviral integration and DNA transposition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7850.map.gz emd_7850.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7850-v30.xml emd-7850-v30.xml emd-7850.xml emd-7850.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7850.png emd_7850.png | 86.8 KB | ||
| Filedesc metadata |  emd-7850.cif.gz emd-7850.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7850 http://ftp.pdbj.org/pub/emdb/structures/EMD-7850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7850 | HTTPS FTP |
-Related structure data
| Related structure data |  6dbuMC  7843C  7844C  7845C  7846C  7847C  7848C  7849C  7851C  7852C  7853C  6dbiC  6dbjC  6dblC  6dboC  6dbqC  6dbrC  6dbtC  6dbvC  6dbwC  6dbxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7850.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7850.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RAG in complex with 12-RSS and 23-RSS substrate DNAs, sharpened map with B factor -190 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
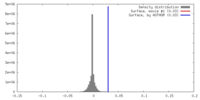
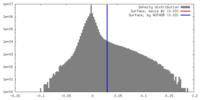
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RAG in complex with 12-RSS and 23-RSS substrate DNAs
| Entire | Name: RAG in complex with 12-RSS and 23-RSS substrate DNAs |
|---|---|
| Components |
|
-Supramolecule #1: RAG in complex with 12-RSS and 23-RSS substrate DNAs
| Supramolecule | Name: RAG in complex with 12-RSS and 23-RSS substrate DNAs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Recombination activating gene 1 - MBP chimera
| Macromolecule | Name: Recombination activating gene 1 - MBP chimera / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 131.160047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH GTKTEEGKLV IWINGDKGYN GLAEVGKKFE KDTGIKVTVE HPDKLEEKFP QVAATGDGPD IIFWAHDRFG GYAQSGLLA EITPDKAFQD KLYPFTWDAV RYNGKLIAYP IAVEALSLIY NKDLLPNPPK TWEEIPALDK ELKAKGKSAL M FNLQEPYF ...String: MGSSHHHHHH GTKTEEGKLV IWINGDKGYN GLAEVGKKFE KDTGIKVTVE HPDKLEEKFP QVAATGDGPD IIFWAHDRFG GYAQSGLLA EITPDKAFQD KLYPFTWDAV RYNGKLIAYP IAVEALSLIY NKDLLPNPPK TWEEIPALDK ELKAKGKSAL M FNLQEPYF TWPLIAADGG YAFKYENGKY DIKDVGVDNA GAKAGLTFLV DLIKNKHMNA DTDYSIAEAA FNKGETAMTI NG PWAWSNI DTSKVNYGVT VLPTFKGQPS KPFVGVLSAG INAASPNKEL AKEFLENYLL TDEGLEAVNK DKPLGAVALK SYE EELAKD PRIAATMENA QKGEIMPNIP QMSAFWYAVR TAVINAASGR QTVDEALKDA QTGTDYDIPT TLEVLFQGPL GSRC QRDHL STKLIPTEVP ADLIRAVTCQ VCDHLLSDPV QSPCRHLFCR LCIIRYTHAL GPNCPTCNQH LNPSHLIKPA KFFLA TLSS LPLLCPSEEC SDWVRLDSFR EHCLNHYREK ESQEEQTPSE QNLDGYLPVN KGGRPRQHLL SLTRRAQKHR LRDLKN QVK TFAEKEEGGD VKSVCLTLFL LALRAGNEHK QADELEAMMQ GRGFGLHPAV CLAIRVNTFL SCSQYHKMYR TVKATSG RQ IFQPLHTLRN AEKELLPGFH QFEWQPALKN VSTSWDVGII DGLSGWTVSV DDVPADTISR RFRYDVALVS ALKDLEED I MEGLRERALD DSMCTSGFTV VVKESCDGMG DVSEKHGSGP AVPEKAVRFS FTIMSISIRL EGEDDGITIF QEQKPNSEL SCRPLCLMFV DESDHETLTA ILGPVVAERK AMMESRLIIS VGGLLRSFRF FFRGTGYDEK MVREMEGLEA SGSTYICTLC DSTRAEASQ NMVLHSITRS HDENLERYEI WRKNPFSESA DELRDRVKGV SAKPFMETQP TLDALHCDIG NATEFYKIFQ D EIGEVYQK PNPSREERRR WRSTLDKQLR KKMKLKPVMR MNGNYARRLM TREAVEAVCE LVPSEERREA LLKLMDLYLQ MK PVWRSTC PSRDCPDQLC QYSYNSQQFA DLLSSMFKYR YDGKITNYLH KTLAHVPEIV ERDGSIGAWA SEGNESGNKL FRR FRKMNA RQSKTFELED ILKHHWLYTS KYLQKFMEAH KNS UniProtKB: Maltose/maltodextrin-binding periplasmic protein, V(D)J recombination-activating protein 1 |
-Macromolecule #2: Recombination activating gene 2
| Macromolecule | Name: Recombination activating gene 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.43593 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGSMSLQPLT AVNCGSLVQP GFSLLDLEGD VYLFGQKGWP KRSCPTGIFG VRIKKGELKL RAISFSNNSS YLPPLRCPAI AHFEAQDGK PECYLIHGGR TPNNELSSSL YMLSVDSRGC NRKVTLRCEE KELVGDVPSA RYGHTLSVIN SRGKTACVLF G GRSYMPPT ...String: GGSMSLQPLT AVNCGSLVQP GFSLLDLEGD VYLFGQKGWP KRSCPTGIFG VRIKKGELKL RAISFSNNSS YLPPLRCPAI AHFEAQDGK PECYLIHGGR TPNNELSSSL YMLSVDSRGC NRKVTLRCEE KELVGDVPSA RYGHTLSVIN SRGKTACVLF G GRSYMPPT ERTTQNWNSV VDCPPQVYLI DLEFGCCTAH TLPELTDGQS FHVALARQDC VYFLGGHILS SDCRPSRLIR LH VELLLGS PVLTCTILHE GLTITSAIAS PIGYHEYIIF GGYQSETQKR MECTYVGLDD VGVHMESREP PQWTSEISHS RTW FGGSLG KGTALVAIPS EGNPTPPEAY HFYQVSFQKE QDGEATAQGG SQESTDFEDS APLEDSEELY FGREPHELEY SSDV EGDTY NEEDEEDESQ TGYWIKCCLS CQVDPNIWEP YYSTELTRPA MIFCSRGEGG HWVHAQCMEL PESLLLQLSQ DNSKY FCLD HGGLPKQEMT PPKQMLPVKR VPMKMTHRKA PVSLKMTPAK KTFLRRLFD UniProtKB: V(D)J recombination-activating protein 2 |
-Macromolecule #3: Forward strand RSS substrate DNA
| Macromolecule | Name: Forward strand RSS substrate DNA / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.450713 KDa |
| Sequence | String: (DG)(DA)(DT)(DC)(DT)(DG)(DG)(DC)(DC)(DT) (DG)(DT)(DC)(DT)(DT)(DA)(DC)(DA)(DC)(DA) (DG)(DT)(DG)(DC)(DT)(DA)(DC)(DA)(DG) (DA)(DC)(DT)(DG)(DG) |
-Macromolecule #4: Reverse strand RSS substrate DNA
| Macromolecule | Name: Reverse strand RSS substrate DNA / type: dna / ID: 4 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.468741 KDa |
| Sequence | String: (DC)(DC)(DA)(DG)(DT)(DC)(DT)(DG)(DT)(DA) (DG)(DC)(DA)(DC)(DT)(DG)(DT)(DG)(DT)(DA) (DA)(DG)(DA)(DC)(DA)(DG)(DG)(DC)(DC) (DA)(DG)(DA)(DT)(DC) |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.38 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Solutions were made fresh from concentrated to avoid microbial contamination. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE | |||||||||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 71.49 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 67194 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)