+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7793 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
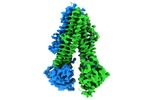
| Title | Thermostablilized dephosphorylated chicken CFTR | |||||||||
 Map data Map data | Sharpened SuperRes PPase-treated chTS map w/o filter by local resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CFTR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRHO GTPases regulate CFTR trafficking / RHOQ GTPase cycle / ABC-family proteins mediated transport / Cargo recognition for clathrin-mediated endocytosis / Clathrin-mediated endocytosis / Aggrephagy / Ub-specific processing proteases / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / bicarbonate transport ...RHO GTPases regulate CFTR trafficking / RHOQ GTPase cycle / ABC-family proteins mediated transport / Cargo recognition for clathrin-mediated endocytosis / Clathrin-mediated endocytosis / Aggrephagy / Ub-specific processing proteases / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / bicarbonate transport / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / chloride channel complex / isomerase activity / chloride transmembrane transport / transmembrane transport / recycling endosome membrane / early endosome membrane / apical plasma membrane / endoplasmic reticulum membrane / ATP binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
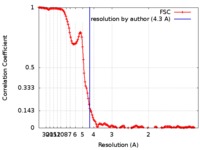
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Fay JF / Riordan JR | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2018 Journal: Biochemistry / Year: 2018Title: Cryo-EM Visualization of an Active High Open Probability CFTR Anion Channel. Authors: Jonathan F Fay / Luba A Aleksandrov / Timothy J Jensen / Liying L Cui / Joseph N Kousouros / Lihua He / Andrei A Aleksandrov / Drew S Gingerich / John R Riordan / James Z Chen /  Abstract: The cystic fibrosis transmembrane conductance regulator (CFTR) anion channel, crucial to epithelial salt and water homeostasis, and defective due to mutations in its gene in patients with cystic ...The cystic fibrosis transmembrane conductance regulator (CFTR) anion channel, crucial to epithelial salt and water homeostasis, and defective due to mutations in its gene in patients with cystic fibrosis, is a unique member of the large family of ATP-binding cassette transport proteins. Regulation of CFTR channel activity is stringently controlled by phosphorylation and nucleotide binding. Structural changes that underlie transitions between active and inactive functional states are not yet fully understood. Indeed the first 3D structures of dephosphorylated, ATP-free, and phosphorylated ATP-bound states were only recently reported. Here we have determined the structure of inactive and active states of a thermally stabilized CFTR, the latter with a very high channel open probability, confirmed after reconstitution into proteoliposomes. These structures, obtained at nominal resolution of 4.3 and 6.6 Å, reveal a unique repositioning of the transmembrane helices and regulatory domain density that provide insights into the structural transition between active and inactive functional states of CFTR. Moreover, we observe an extracellular vestibule that may provide anion access to the pore due to the conformation of transmembrane helices 7 and 8 that differs from the previous orthologue CFTR structures. In conclusion, our work contributes detailed structural information on an active, open state of the CFTR anion channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7793.map.gz emd_7793.map.gz | 227.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7793-v30.xml emd-7793-v30.xml emd-7793.xml emd-7793.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7793_fsc.xml emd_7793_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_7793.png emd_7793.png | 114.3 KB | ||
| Masks |  emd_7793_msk_1.map emd_7793_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-7793.cif.gz emd-7793.cif.gz | 6.2 KB | ||
| Others |  emd_7793_additional_1.map.gz emd_7793_additional_1.map.gz emd_7793_additional_2.map.gz emd_7793_additional_2.map.gz emd_7793_half_map_1.map.gz emd_7793_half_map_1.map.gz emd_7793_half_map_2.map.gz emd_7793_half_map_2.map.gz | 12.8 MB 28.6 MB 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7793 http://ftp.pdbj.org/pub/emdb/structures/EMD-7793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7793 | HTTPS FTP |
-Validation report
| Summary document |  emd_7793_validation.pdf.gz emd_7793_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7793_full_validation.pdf.gz emd_7793_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_7793_validation.xml.gz emd_7793_validation.xml.gz | 22.4 KB | Display | |
| Data in CIF |  emd_7793_validation.cif.gz emd_7793_validation.cif.gz | 28.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7793 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7793 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7793 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7793 | HTTPS FTP |
-Related structure data
| Related structure data |  6d3rMC  7794C  6d3sC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10219 (Title: Cryo-electron microscopy data of thermostabilized avian CFTR EMPIAR-10219 (Title: Cryo-electron microscopy data of thermostabilized avian CFTRData size: 19.3 Data #1: Binned Particle stacks and meta data for cryo-EM structures of phosphorylated and dephosphorylated avian CFTR [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7793.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7793.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened SuperRes PPase-treated chTS map w/o filter by local resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68866 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
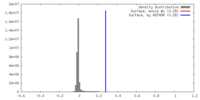
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7793_msk_1.map emd_7793_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
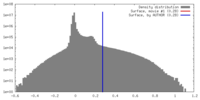
| Density Histograms |
-Additional map: Sharpened SuperRes PPase-treated chTS map w/ filter by...
| File | emd_7793_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened SuperRes PPase-treated chTS map w/ filter by local resolution | ||||||||||||
| Projections & Slices |
| ||||||||||||
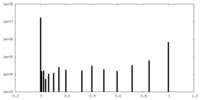
| Density Histograms |
-Additional map: Binned PPase-treated chTS map
| File | emd_7793_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Binned PPase-treated chTS map | ||||||||||||
| Projections & Slices |
| ||||||||||||
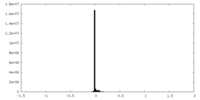
| Density Histograms |
-Half map: raw half map A
| File | emd_7793_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | raw half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: raw half map B
| File | emd_7793_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | raw half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CFTR
| Entire | Name: CFTR |
|---|---|
| Components |
|
-Supramolecule #1: CFTR
| Supramolecule | Name: CFTR / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cystic fibrosis transmembrane conductance regulator
| Macromolecule | Name: Cystic fibrosis transmembrane conductance regulator / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ec: 3.6.3.49 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 162.637438 KDa |
| Recombinant expression | Organism:  Cricetinae (hamsters) Cricetinae (hamsters) |
| Sequence | String: MQRSPLEKAN IFSKLFFRWT KPILKKGYRQ RLELSDIYQI PSADSADNLS EKLEREWDRE LATSKKKPKL INALRRCFFW KFMFYGILL YLGEVTKSVQ PLLLGRIIAS YDPDNSSERS IAYYLGIGLC LLFLVRTLLI HPSIFGLHHI GMQIRIALFS L IYKKTLKL ...String: MQRSPLEKAN IFSKLFFRWT KPILKKGYRQ RLELSDIYQI PSADSADNLS EKLEREWDRE LATSKKKPKL INALRRCFFW KFMFYGILL YLGEVTKSVQ PLLLGRIIAS YDPDNSSERS IAYYLGIGLC LLFLVRTLLI HPSIFGLHHI GMQIRIALFS L IYKKTLKL SSKVLDKIST GQLVSLLSNN LNKFDEGLAL AHFVWIAPLQ VALLMGLLWD MLQASAFAGL AFLIVMAFFQ AW LGQMMMK YRDKRAGKIN ERLVITSEII ENIQSVKAYC WEDAMEKMIE SLRETELKLT RKAAYVRYFN SSAFFFSGFF VVF LAVVPY AVTKGIILRK IFTTISFCIV LRMTVTRQFP GSVQTWYDSI GAINKIQDFL LKEEYKALEY NLTTTGVEVD KVTA FWDEH ASPVLQDINF KIEKGELLAV SGSTGSGKTS LLMLIMGELE PSEGKIKHSG RISFSPQVSW IMPGTIKENI IFGVS YDEY RYKSVIQACQ LEEDILKFPD KDYTVLGEGG IILSGGQRAR ISLARAVYKD ADLYLMDSPF GYLDIFTEKE IFESCV CKL MANKTRILVT SKLEHLKIAD KILILHEGSC YFYGTFSELQ GQRPDFSSEL MGFDSFDQFS AERRNSIITE TLRRFSF EG ESMGSRNEMK KQSFKQTSDF NDKRKNSIII NPLNAGRKLS IMQKNGTQVN GLEDGHIDSP ERRISLVPDL EQGDVGLP R SNMLNSDHML QSRRRQSVLS LMTGTSVNQG PHVSKKGSTS FRKMSVVPQT NLSSEIDIYT RRLSRDSILD ITDEINEED LKECFTDDAE SMGTVTTWNT YFRYITIHKS LIFVLILCVT IFLLEVAASL VLLLFLQKAA QINATQPENA TSDNPPVIIT DTSSYYMIY IYVGIADTLL AMGIFRGLPL VHTLITVSKT LHQKMVHAVL YAPMSTFNSL KAGGILNRFS KDTAILDDLL P LTVFDLIQ LILIVIGAIT VVSILQPYIF LASVPVIAAF IVLRAYFLHT SQQLKQLESE ARSPIFTHLV TSLKGLWTLR AF GRQPYFE TLFHKALNLH TANWFLYLST LRWFQMRIEM IFVVFFSAVA FISIITTGDG PGRVGIILTL AMNIMGTLQW AVN SSIDVD SLMRSVSRIF KFIDMPTEEM KTIKPQKNNQ FSDALIIENR HVKDEKNWPS GGQMTVTDLT ARYTEGGTAV LENI SFSIS SGQTVGLLGR TGSGKSTLLF AFLRLLNTEG DIQIDGVSWN TVSLQQWRKA FGVIPQKVFI FSGTFRKNLD PYGQW NDEE IWKVAEEVGL KSVIEQFPGQ LDFVLVDGGC VLSHGHKQLM CLARSVLSKA KILLLDEPSA HLDPITSQVI RKTLKH AFA DCTVVLSESR LEAILECQRF LVIEDNKMRQ YESIQKLLSE KSSLRQSGSG GGGGGSLEVL FQGDHHHHHH HHHH |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)