+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7477 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cdc48 in the presence of ADP | |||||||||
 Map data Map data | Cdc48-Npl4 complex in the presence of ATP-gamma-S | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | |||||||||
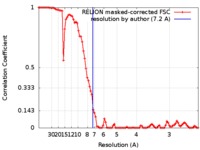
| Method | single particle reconstruction / cryo EM / Resolution: 7.2 Å | |||||||||
 Authors Authors | Kim KH / Bodnar NO | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4. Authors: Nicholas O Bodnar / Kelly H Kim / Zhejian Ji / Thomas E Wales / Vladimir Svetlov / Evgeny Nudler / John R Engen / Thomas Walz / Tom A Rapoport /  Abstract: Many polyubiquitinated proteins are extracted from membranes or complexes by the conserved ATPase Cdc48 (in yeast; p97 or VCP in mammals) before proteasomal degradation. Each Cdc48 hexamer contains ...Many polyubiquitinated proteins are extracted from membranes or complexes by the conserved ATPase Cdc48 (in yeast; p97 or VCP in mammals) before proteasomal degradation. Each Cdc48 hexamer contains two stacked ATPase rings (D1 and D2) and six N-terminal (N) domains. Cdc48 binds various cofactors, including the Ufd1-Npl4 heterodimer. Here, we report structures of the Cdc48-Ufd1-Npl4 complex from Chaetomium thermophilum. Npl4 interacts through its UBX-like domain with a Cdc48 N domain, and it uses two Zn-finger domains to anchor the enzymatically inactive Mpr1-Pad1 N-terminal (MPN) domain, homologous to domains found in several isopeptidases, to the top of the D1 ATPase ring. The MPN domain of Npl4 is located above Cdc48's central pore, a position similar to the MPN domain from deubiquitinase Rpn11 in the proteasome. Our results indicate that Npl4 is unique among Cdc48 cofactors and suggest a mechanism for binding and translocation of polyubiquitinated substrates into the ATPase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7477.map.gz emd_7477.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7477-v30.xml emd-7477-v30.xml emd-7477.xml emd-7477.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7477_fsc.xml emd_7477_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_7477.png emd_7477.png | 95.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7477 http://ftp.pdbj.org/pub/emdb/structures/EMD-7477 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7477 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7477 | HTTPS FTP |
-Validation report
| Summary document |  emd_7477_validation.pdf.gz emd_7477_validation.pdf.gz | 78.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7477_full_validation.pdf.gz emd_7477_full_validation.pdf.gz | 77.3 KB | Display | |
| Data in XML |  emd_7477_validation.xml.gz emd_7477_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7477 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7477 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7477 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7477 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7477.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7477.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cdc48-Npl4 complex in the presence of ATP-gamma-S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cdc48
| Entire | Name: Cdc48 |
|---|---|
| Components |
|
-Supramolecule #1: Cdc48
| Supramolecule | Name: Cdc48 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Cdc48
| Macromolecule | Name: Cdc48 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Recombinant expression | Organism:  |
| Sequence | String: MAPESKDHKK KVNLTDPSGA EHREENDTAT AILKKKKKPN QLMVTDAVND DNSIIALSNN TMEALQLFRG DTVLVRGKKR RDTVLIVLAD DDLDDGSARI NRVVRHNLRV KHGDMITIHP CPDIKYAKRI AVLPIADTVE GLTGSLFDVF LAPYFREAYR PVRQGDLFTV ...String: MAPESKDHKK KVNLTDPSGA EHREENDTAT AILKKKKKPN QLMVTDAVND DNSIIALSNN TMEALQLFRG DTVLVRGKKR RDTVLIVLAD DDLDDGSARI NRVVRHNLRV KHGDMITIHP CPDIKYAKRI AVLPIADTVE GLTGSLFDVF LAPYFREAYR PVRQGDLFTV RGGMRQVEFK VVEVDPPEYG IVAQDTIIHC EGEPIPREEE ENNLNEVGYD DIGGCRKQLA QIREMVELPL RHPQLFKSIG IKPPRGVLLY GPPGTGKTLM ARAVANETGA FFFLINGPEI MSKMAGESES NLRKAFEEAE KNSPAIIFID EIDSIAPKRE KTNGEVERRV VSQLLTLMDG MKARSNVVVM AATNRPNSID PALRRFGRFD REVDIGIPDP TGRLEILQIH TKNMKLADDV DLEQIAAETH GYVGSDLAAL CSEAAMQQIR EKMDLIDLDE DTIDAEVLDS LGVTMDNFRY ALGVSNPSAL REVAVVEVPN VRWEDIGGLE QVKQELKEQV QYPVDHPEKF LKFGLSPSRG VLFYGPPGTG KTMLAKAVAN ECAANFISVK GPELLSMWFG ESESNIRDIF DKARAAAPCV VFLDELDSIA KARGGSIGDA GGASDRVVNQ LLTEMDGMTS KKNVFVIGAT NRPEQLDPAL CRPGRLDQLI YVPLPDEAGR LSILKAQLRK TPVSKDVDLA YIASKTHGFS GADLAFITQR AVKLAIKESI AAEIERQKAR EAAGEDVNME DDEDPVPELT KRHFEEAMRD ARRSVSDVEI RRYEAFAQQM KNAGPGAFFK FPDSTTDNSA SNAAGNSFGD AGNDDDLYT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 1628 / Average exposure time: 6.0 sec. / Average electron dose: 1.04 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 31000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)