[English] 日本語
 Yorodumi
Yorodumi- EMDB-7339: Cryo-EM structure of human KATP bound to ATP and ADP in propeller form -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7339 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human KATP bound to ATP and ADP in propeller form | |||||||||
 Map data Map data | human KATP complexed with ATP and ADP in propeller form | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP-dependent potassium channel Kir6.2 SUR1 ATP ADP / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective ABCC9 causes CMD10, ATFB12 and Cantu syndrome / negative regulation of neuroblast migration / positive regulation of uterine smooth muscle relaxation / ATP sensitive Potassium channels / Defective ABCC8 can cause hypo- and hyper-glycemias / potassium ion-transporting ATPase complex / negative regulation of blood-brain barrier permeability / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / response to resveratrol ...Defective ABCC9 causes CMD10, ATFB12 and Cantu syndrome / negative regulation of neuroblast migration / positive regulation of uterine smooth muscle relaxation / ATP sensitive Potassium channels / Defective ABCC8 can cause hypo- and hyper-glycemias / potassium ion-transporting ATPase complex / negative regulation of blood-brain barrier permeability / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / response to resveratrol / inward rectifying potassium channel / sulfonylurea receptor activity / ventricular cardiac muscle tissue development / positive regulation of tight junction disassembly / CAMKK-AMPK signaling cascade / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / regulation of monoatomic ion transmembrane transport / positive regulation of potassium ion transport / ATPase-coupled monoatomic cation transmembrane transporter activity / response to pH / negative regulation of low-density lipoprotein particle clearance / nervous system process / : / ankyrin binding / negative regulation of glial cell proliferation / response to zinc ion / neuromuscular process / response to ATP / potassium ion import across plasma membrane / intracellular glucose homeostasis / potassium ion binding / action potential / voltage-gated potassium channel activity / ATPase-coupled transmembrane transporter activity / potassium channel activity / ABC-type transporter activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / Ion homeostasis / potassium ion transmembrane transport / cellular response to nutrient levels / T-tubule / regulation of insulin secretion / negative regulation of angiogenesis / response to ischemia / determination of adult lifespan / regulation of membrane potential / potassium ion transport / Regulation of insulin secretion / female pregnancy / negative regulation of insulin secretion / ADP binding / sarcolemma / visual learning / response to insulin / ABC-family proteins mediated transport / transmembrane transport / memory / glucose metabolic process / positive regulation of tumor necrosis factor production / synaptic vesicle membrane / presynaptic membrane / response to lipopolysaccharide / transmembrane transporter binding / response to hypoxia / response to xenobiotic stimulus / apoptotic process / ATP hydrolysis activity / ATP binding / metal ion binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||
 Authors Authors | Lee KPK / Chen J | |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Molecular structure of human KATP in complex with ATP and ADP. Authors: Kenneth Pak Kin Lee / Jue Chen / Roderick MacKinnon /  Abstract: In many excitable cells, KATP channels respond to intracellular adenosine nucleotides: ATP inhibits while ADP activates. We present two structures of the human pancreatic KATP channel, containing the ...In many excitable cells, KATP channels respond to intracellular adenosine nucleotides: ATP inhibits while ADP activates. We present two structures of the human pancreatic KATP channel, containing the ABC transporter SUR1 and the inward-rectifier K channel Kir6.2, in the presence of Mg and nucleotides. These structures, referred to as quatrefoil and propeller forms, were determined by single-particle cryo-EM at 3.9 Å and 5.6 Å, respectively. In both forms, ATP occupies the inhibitory site in Kir6.2. The nucleotide-binding domains of SUR1 are dimerized with Mg-ATP in the degenerate site and Mg-ADP in the consensus site. A lasso extension forms an interface between SUR1 and Kir6.2 adjacent to the ATP site in the propeller form and is disrupted in the quatrefoil form. These structures support the role of SUR1 as an ADP sensor and highlight the lasso extension as a key regulatory element in ADP's ability to override ATP inhibition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7339.map.gz emd_7339.map.gz | 95.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7339-v30.xml emd-7339-v30.xml emd-7339.xml emd-7339.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7339.png emd_7339.png | 83.7 KB | ||
| Filedesc metadata |  emd-7339.cif.gz emd-7339.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7339 http://ftp.pdbj.org/pub/emdb/structures/EMD-7339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7339 | HTTPS FTP |
-Related structure data
| Related structure data |  6c3pMC  7338C  6c3oC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7339.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7339.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human KATP complexed with ATP and ADP in propeller form | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
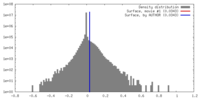
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Quatrefoil form of human KATP in complex with ATP and ADP
| Entire | Name: Quatrefoil form of human KATP in complex with ATP and ADP |
|---|---|
| Components |
|
-Supramolecule #1: Quatrefoil form of human KATP in complex with ATP and ADP
| Supramolecule | Name: Quatrefoil form of human KATP in complex with ATP and ADP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 880 KDa |
-Macromolecule #1: ATP-sensitive inward rectifier potassium channel 11
| Macromolecule | Name: ATP-sensitive inward rectifier potassium channel 11 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.144367 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SASASAMLSR KGIIPEEYVL TRLAEDPAEP RYRARQRRAR FVSKKGNCNV AHKNIREQGR FLQDVFTTLV DLKWPHTLLI FTMSFLCSW LLFAMAWWLI AFAHGDLAPS EGTAEPCVTS IHSFSSAFLF SIEVQVTIGF GGRMVTEECP LAILILIVQN I VGLMINAI ...String: SASASAMLSR KGIIPEEYVL TRLAEDPAEP RYRARQRRAR FVSKKGNCNV AHKNIREQGR FLQDVFTTLV DLKWPHTLLI FTMSFLCSW LLFAMAWWLI AFAHGDLAPS EGTAEPCVTS IHSFSSAFLF SIEVQVTIGF GGRMVTEECP LAILILIVQN I VGLMINAI MLGCIFMKTA QAHRRAETLI FSKHAVIALR HGRLCFMLRV GDLRKSMIIS ATIHMQVVRK TTSPEGEVVP LH QVDIPME NGVGGNSIFL VAPLIIYHVI DANSPLYDLA PSDLHHHQDL EIIVILEGVV ETTGITTQAR TSYLADEILW GQR FVPIVA EEDGRYSVDY SKFGNTIKVP TPLCTARQLD EDHSLLEALT LASARGPLRK RSVPMAKAKP KFSISPDSLS SNSL EVLFQ G UniProtKB: ATP-sensitive inward rectifier potassium channel 11 |
-Macromolecule #2: ATP-binding cassette sub-family C member 8
| Macromolecule | Name: ATP-binding cassette sub-family C member 8 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 177.180219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPLAFCGSEN HSAAYRVDQG VLNNGCFVDA LNVVPHVFLL FITFPILFIG WGSQSSKVHI HHSTWLHFPG HNLRWILTFM LLFVLVCEI AEGILSDGVT ESHHLHLYMP AGMAFMAAVT SVVYYHNIET SNFPKLLIAL LVYWTLAFIT KTIKFVKFLD H AIGFSQLR ...String: MPLAFCGSEN HSAAYRVDQG VLNNGCFVDA LNVVPHVFLL FITFPILFIG WGSQSSKVHI HHSTWLHFPG HNLRWILTFM LLFVLVCEI AEGILSDGVT ESHHLHLYMP AGMAFMAAVT SVVYYHNIET SNFPKLLIAL LVYWTLAFIT KTIKFVKFLD H AIGFSQLR FCLTGLLVIL YGMLLLVEVN VIRVRRYIFF KTPREVKPPE DLQDLGVRFL QPFVNLLSKG TYWWMNAFIK TA HKKPIDL RAIGKLPIAM RALTNYQRLC EAFDAQVRKD IQGTQGARAI WQALSHAFGR RLVLSSTFRI LADLLGFAGP LCI FGIVDH LGKENDVFQP KTQFLGVYFV SSQEFLANAY VLAVLLFLAL LLQRTFLQAS YYVAIETGIN LRGAIQTKIY NKIM HLSTS NLSMGEMTAG QICNLVAIDT NQLMWFFFLC PNLWAMPVQI IVGVILLYYI LGVSALIGAA VIILLAPVQY FVATK LSQA QRSTLEYSNE RLKQTNEMLR GIKLLKLYAW ENIFRTRVET TRRKEMTSLR AFAIYTSISI FMNTAIPIAA VLITFV GHV SFFKEADFSP SVAFASLSLF HILVTPLFLL SSVVRSTVKA LVSVQKLSEF LSSAEIREEQ CAPHEPTPQG PASKYQA VP LRVVNRKRPA REDCRGLTGP LQSLVPSADG DADNCCVQIM GGYFTWTPDG IPTLSNITIR IPRGQLTMIV GQVGCGKS S LLLAALGEMQ KVSGAVFWSS LPDSEIGEDP SPERETATDL DIRKRGPVAY ASQKPWLLNA TVEENIIFES PFNKQRYKM VIEACSLQPD IDILPHGDQT QIGERGINLS GGQRQRISVA RALYQHANVV FLDDPFSALD IHLSDHLMQA GILELLRDDK RTVVLVTHK LQYLPHADWI IAMKDGTIQR EGTLKDFQRS ECQLFEHWKT LMNRQDQELE KETVTERKAT EPPQGLSRAM S SRDGLLQD EEEEEEEAAE SEEDDNLSSM LHQRAEIPWR ACAKYLSSAG ILLLSLLVFS QLLKHMVLVA IDYWLAKWTD SA LTLTPAA RNCSLSQECT LDQTVYAMVF TVLCSLGIVL CLVTSVTVEW TGLKVAKRLH RSLLNRIILA PMRFFETTPL GSI LNRFSS DCNTIDQHIP STLECLSRST LLCVSALAVI SYVTPVFLVA LLPLAIVCYF IQKYFRVASR DLQQLDDTTQ LPLL SHFAE TVEGLTTIRA FRYEARFQQK LLEYTDSNNI ASLFLTAANR WLEVRMEYIG ACVVLIAAVT SISNSLHREL SAGLV GLGL TYALMVSNYL NWMVRNLADM ELQLGAVKRI HGLLKTEAES YEGLLAPSLI PKNWPDQGKI QIQNLSVRYD SSLKPV LKH VNALIAPGQK IGICGRTGSG KSSFSLAFFR MVDTFEGHII IDGIDIAKLP LHTLRSRLSI ILQDPVLFSG TIRFNLD PE RKCSDSTLWE ALEIAQLKLV VKALPGGLDA IITEGGENFS QGQRQLFCLA RAFVRKTSIF IMDEATASID MATENILQ K VVMTAFADRT VVTIAHRVHT ILSADLVIVL KRGAILEFDK PEKLLSRKDS VFASFVRADK UniProtKB: ATP-binding cassette sub-family C member 8 |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 8 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 8 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.45 mg/mL |
|---|---|
| Buffer | pH: 8.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.18 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL / In silico model: ab-initio model from cryoSPARC |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 47282 |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: cryoSPARC |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)