+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7101 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | TPX2_mini decorated GMPCPP-microtubule | ||||||||||||||||||
 Map data Map data | TPX2_mini decorated GMPCPP- microtubule | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | mitosis / GanGTP / nucleation / CELL CYCLE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationactivation of protein kinase activity / axon hillock / negative regulation of microtubule depolymerization / microtubule nucleation / importin-alpha family protein binding / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport ...activation of protein kinase activity / axon hillock / negative regulation of microtubule depolymerization / microtubule nucleation / importin-alpha family protein binding / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / COPI-mediated anterograde transport / mitotic spindle assembly / intercellular bridge / AURKA Activation by TPX2 / regulation of mitotic spindle organization / protein serine/threonine kinase activator activity / structural constituent of cytoskeleton / microtubule cytoskeleton organization / spindle / neuron migration / spindle pole / mitotic spindle / mitotic cell cycle / microtubule cytoskeleton / Regulation of TP53 Activity through Phosphorylation / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / molecular adaptor activity / microtubule / cell division / GTPase activity / apoptotic process / protein kinase binding / GTP binding / nucleoplasm / metal ion binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||
 Authors Authors | Zhang R / Nogales E | ||||||||||||||||||
| Funding support |  United States, United States,  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Structural insight into TPX2-stimulated microtubule assembly. Authors: Rui Zhang / Johanna Roostalu / Thomas Surrey / Eva Nogales /   Abstract: During mitosis and meiosis, microtubule (MT) assembly is locally upregulated by the chromatin-dependent Ran-GTP pathway. One of its key targets is the MT-associated spindle assembly factor TPX2. The ...During mitosis and meiosis, microtubule (MT) assembly is locally upregulated by the chromatin-dependent Ran-GTP pathway. One of its key targets is the MT-associated spindle assembly factor TPX2. The molecular mechanism of how TPX2 stimulates MT assembly remains unknown because structural information about the interaction of TPX2 with MTs is lacking. Here, we determine the cryo-electron microscopy structure of a central region of TPX2 bound to the MT surface. TPX2 uses two flexibly linked elements ('ridge' and 'wedge') in a novel interaction mode to simultaneously bind across longitudinal and lateral tubulin interfaces. These MT-interacting elements overlap with the binding site of importins on TPX2. Fluorescence microscopy-based in vitro reconstitution assays reveal that this interaction mode is critical for MT binding and facilitates MT nucleation. Together, our results suggest a molecular mechanism of how the Ran-GTP gradient can regulate TPX2-dependent MT formation. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7101.map.gz emd_7101.map.gz | 155.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7101-v30.xml emd-7101-v30.xml emd-7101.xml emd-7101.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7101.png emd_7101.png | 408.4 KB | ||
| Filedesc metadata |  emd-7101.cif.gz emd-7101.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7101 http://ftp.pdbj.org/pub/emdb/structures/EMD-7101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7101 | HTTPS FTP |
-Related structure data
| Related structure data |  6bjcMC  7102C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7101.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7101.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TPX2_mini decorated GMPCPP- microtubule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
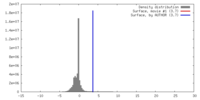
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TPX2_mini decorated GMPCPP-microtubule
| Entire | Name: TPX2_mini decorated GMPCPP-microtubule |
|---|---|
| Components |
|
-Supramolecule #1: TPX2_mini decorated GMPCPP-microtubule
| Supramolecule | Name: TPX2_mini decorated GMPCPP-microtubule / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Tubulin
| Supramolecule | Name: Tubulin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Targeting protein for Xklp2
| Supramolecule | Name: Targeting protein for Xklp2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.204445 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRSIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GVDSVEGEGE EEGEEY UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.90777 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAVF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATADEQGE FEEEGEEDEA UniProtKB: Tubulin beta chain |
-Macromolecule #3: Targeting protein for Xklp2
| Macromolecule | Name: Targeting protein for Xklp2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 85.811328 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) |
| Sequence | String: MSQVKSSYSY DAPSDFINFS SLDDEGDTQN IDSWFEEKAN LENKLLGKNG TGGLFQGKTP LRKANLQQAI VTPLKPVDNT YYKEAEKEN LVEQSIPSNA CSSLEVEAAI SRKTPAQPQR RSLRLSAQKD LEQKEKHHVK MKAKRCATPV IIDEILPSKK M KVSNNKKK ...String: MSQVKSSYSY DAPSDFINFS SLDDEGDTQN IDSWFEEKAN LENKLLGKNG TGGLFQGKTP LRKANLQQAI VTPLKPVDNT YYKEAEKEN LVEQSIPSNA CSSLEVEAAI SRKTPAQPQR RSLRLSAQKD LEQKEKHHVK MKAKRCATPV IIDEILPSKK M KVSNNKKK PEEEGSAHQD TAEKNASSPE KAKGRHTVPC MPPAKQKFLK STEEQELEKS MKMQQEVVEM RKKNEEFKKL AL AGIGQPV KKSVSQVTKS VDFHFRTDER IKQHPKNQEE YKEVNFTSEL RKHPSSPARV TKGCTIVKPF NLSQGKKRTF DET VSTYVP LAQQVEDFHK RTPNRYHLRS KKDDINLLPS KSSVTKICRD PQTPVLQTKH RARAVTCKST AELEAEELEK LQQY KFKAR ELDPRILEGG PILPKKPPVK PPTEPIGFDL EIEKRIQERE SKKKTEDEHF EFHSRPCPTK ILEDVVGVPE KKVLP ITVP KSPAFALKNR IRMPTKEDEE EDEPVVIKAQ PVPHYGVPFK PQIPEARTVE ICPFSFDSRD KERQLQKEKK IKELQK GEV PKFKALPLPH FDTINLPEKK VKNVTQIEPF CLETDRRGAL KAQTWKHQLE EELRQQKEAA CFKARPNTVI SQEPFVP KK EKKSVAEGLS GSLVQEPFQL ATEKRAKERQ ELEKRMAEVE AQKAQQLEEA RLQEEEQKKE ELARLRRELV HKANPIRK Y QGLEIKSSDQ PLTVPVSPKF STRFHC UniProtKB: Targeting protein for Xklp2 |
-Macromolecule #4: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 6 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 12 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 6 / Number of copies: 6 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 / Details: BRB80 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK II / Details: blot for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 4 / Number real images: 2000 / Average exposure time: 6.0 sec. / Average electron dose: 27.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 9.03 Å Applied symmetry - Helical parameters - Δ&Phi: -25.74 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN / Number images used: 85000 |
|---|---|
| Startup model | Type of model: EMDB MAP |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: FREALIGN |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6bjc: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)