[English] 日本語
 Yorodumi
Yorodumi- EMDB-7099: Cryo-EM structure of ATP-bound, outward-facing bovine multidrug r... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7099 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP-bound, outward-facing bovine multidrug resistance protein 1 (MRP1) | ||||||||||||
 Map data Map data | Full map scaled to model, B-factor sharpened with a sharpening factor of -75 A^2. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ABC transporter / multidrug resistance / outward facing / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationSphingolipid de novo biosynthesis / Heme degradation / Synthesis of Leukotrienes (LT) and Eoxins (EX) / Transport of RCbl within the body / cyclic nucleotide transport / Paracetamol ADME / ABC-family proteins mediated transport / glutathione transmembrane transport / leukotriene transport / ABC-type glutathione-S-conjugate transporter ...Sphingolipid de novo biosynthesis / Heme degradation / Synthesis of Leukotrienes (LT) and Eoxins (EX) / Transport of RCbl within the body / cyclic nucleotide transport / Paracetamol ADME / ABC-family proteins mediated transport / glutathione transmembrane transport / leukotriene transport / ABC-type glutathione-S-conjugate transporter / Cytoprotection by HMOX1 / ABC-type glutathione S-conjugate transporter activity / glutathione transmembrane transporter activity / ABC-type xenobiotic transporter / ABC-type xenobiotic transporter activity / lipid transport / xenobiotic transmembrane transporter activity / xenobiotic transport / ABC-type transporter activity / positive regulation of inflammatory response / basolateral plasma membrane / response to xenobiotic stimulus / ATP hydrolysis activity / ATP binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.14 Å | ||||||||||||
 Authors Authors | Johnson ZL / Chen J | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: ATP Binding Enables Substrate Release from Multidrug Resistance Protein 1. Authors: Zachary Lee Johnson / Jue Chen /  Abstract: The multidrug resistance protein MRP1 is an ATP-driven pump that confers resistance to chemotherapy. Previously, we have shown that intracellular substrates are recruited to a bipartite binding site ...The multidrug resistance protein MRP1 is an ATP-driven pump that confers resistance to chemotherapy. Previously, we have shown that intracellular substrates are recruited to a bipartite binding site when the transporter rests in an inward-facing conformation. A key question remains: how are high-affinity substrates transferred across the membrane and released outside the cell? Using electron cryomicroscopy, we show here that ATP binding opens the transport pathway to the extracellular space and reconfigures the substrate-binding site such that it relinquishes its affinity for substrate. Thus, substrate is released prior to ATP hydrolysis. With this result, we now have a complete description of the conformational cycle that enables substrate transfer in a eukaryotic ABC exporter. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
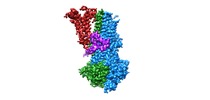
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7099.map.gz emd_7099.map.gz | 201.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7099-v30.xml emd-7099-v30.xml emd-7099.xml emd-7099.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7099.png emd_7099.png | 128.9 KB | ||
| Filedesc metadata |  emd-7099.cif.gz emd-7099.cif.gz | 7.2 KB | ||
| Others |  emd_7099_additional.map.gz emd_7099_additional.map.gz emd_7099_half_map_1.map.gz emd_7099_half_map_1.map.gz emd_7099_half_map_2.map.gz emd_7099_half_map_2.map.gz | 199.6 MB 199.7 MB 199.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7099 http://ftp.pdbj.org/pub/emdb/structures/EMD-7099 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7099 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7099 | HTTPS FTP |
-Related structure data
| Related structure data |  6bhuMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7099.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7099.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map scaled to model, B-factor sharpened with a sharpening factor of -75 A^2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
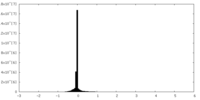
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.817 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
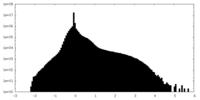
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Full map scaled to model, unsharpened.
| File | emd_7099_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map scaled to model, unsharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
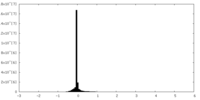
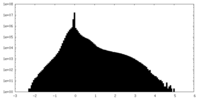
| Density Histograms |
-Half map: Half map 2 scaled to model, unsharpened.
| File | emd_7099_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 scaled to model, unsharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
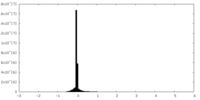
| Density Histograms |
-Half map: Half map 1 scaled to model, unsharpened.
| File | emd_7099_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 scaled to model, unsharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
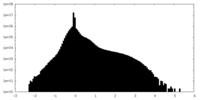
| Density Histograms |
- Sample components
Sample components
-Entire : bovine multidrug resistance protein 1 (MRP1) E1454Q
| Entire | Name: bovine multidrug resistance protein 1 (MRP1) E1454Q |
|---|---|
| Components |
|
-Supramolecule #1: bovine multidrug resistance protein 1 (MRP1) E1454Q
| Supramolecule | Name: bovine multidrug resistance protein 1 (MRP1) E1454Q / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 170 KDa |
-Macromolecule #1: Multidrug resistance-associated protein 1
| Macromolecule | Name: Multidrug resistance-associated protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 183.074062 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)MAL RDFCSVDGSD LFWEWNVTWN TSNPDFTKCF QNTVLVW VP CSYLWVCFPF YFLYLSHHDR GYIQMTHLNK AKTALGFLLW IVCWADLFYS FWERSMGKLL APVFLVSPTL LGITMLLA T FLIQIERRRG VQSSGIMLTF WLIALLCALA ILRSKIMTAL KEDARVDVFR DVTFYIYFSL VLIQLVLSCF SDRSPLFSE TINDPNPCPE SSASFLSRIT FWWITGMMVQ GYRQPLESTD LWSLNKEDTS EQVVPVLVKN WKKECAKSRK QPVKIVYSSK DPAKPKGSS KVDVNEEAEA LIVKCPQKER DPSLFKVLYK TFGPYFLMSF LFKAVHDLMM FAGPEILKLL INFVNDKKAP E WQGYFYTA LLFISACLQT LVLHQYFHIC FVSGMRIKTA VIGAVYRKAL VITNAARKSS TVGEIVNLMS VDAQRFMDLA TY INMIWSA PLQVILALYL LWLNLGPSVL AGVAVMVLMV PLNAVMAMKT KTYQVAHMKS KDNRIKLMNE ILNGIKVLKL YAW ELAFKD KVLAIRQEEL KVLKKSAYLA AVGTFTWVCT PFLVALSTFA VYVTVDENNI LDAQKAFVSL ALFNILRFPL NILP MVISS IVQASVSLKR LRVFLSHEDL DPDSIQRRPI KDAGATNSIT VKNATFTWAR NDPPTLHGIT FSVPEGSLVA VVGQV GCGK SSLLSALLAE MDKVEGHVTV KGSVAYVPQQ AWIQNISLRE NILFGRQLQE RYYKAVVEAC ALLPDLEILP SGDRTE IGE KGVNLSGGQK QRVSLARAVY CDSDVYLLDD PLSAVDAHVG KHIFENVIGP KGLLKNKTRL LVTHAISYLP QMDVIIV MS GGKISEMGSY QELLARDGAF AEFLRTYASA EQEQGQPEDG LAGVGGPGKE VKQMENGMLV TDTAGKQMQR QLSSSSSY S RDVSQHHTST AELRKPGPTE ETWKLVEADK AQTGQVKLSV YWDYMKAIGL FISFLSIFLF LCNHVASLVS NYWLSLWTD DPIVNGTQEH TQVRLSVYGA LGISQGITVF GYSMAVSIGG IFASRRLHLD LLHNVLRSPI SFFERTPSGN LVNRFSKELD TVDSMIPQV IKMFMGSLFN VIGACIIILL ATPMAAVIIP PLGLIYFFVQ RFYVASSRQL KRLESVSRSP VYSHFNETLL G VSVIRAFE EQERFIRQSD LKVDENQKAY YPSIVANRWL AVRLECVGNC IVLFASLFAV ISRHSLSAGL VGLSVSYSLQ VT TYLNWLV RMSSEMETNI VAVERLKEYS ETEKEAPWQI QDMAPPKDWP QVGRVEFRDY GLRYREDLDL VLKHINVTID GGE KVGIVG RTGAGKSSLT LGLFRIKESA EGEIIIDDIN IAKIGLHDLR FKITIIPQDP VLFSGSLRMN LDPFSQYSDE EVWT SLELA HLKGFVSALP DKLNHECAEG GENLSVGQRQ LVCLARALLR KTKILVLDQA TAAVDLETDD LIQSTIRTQF DDCTV LTIA HRLNTIMDYT RVIVLDKGEI QEWGSPSDLL QQRGLFYSMA KDSGLVSNSL EVLFQ UniProtKB: Multidrug resistance-associated protein 1 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 3 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.9 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 400-mesh Au Holey Carbon Grids / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 12 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 2 / Number real images: 4210 / Average exposure time: 7.0 sec. / Average electron dose: 84.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 37000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6bhu: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)