+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7086 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | active dimer | |||||||||
 Map data Map data | Primary consensus dimer map in C2. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PIKK / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of type B pancreatic cell development / RNA polymerase III type 2 promoter sequence-specific DNA binding / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / regulation of locomotor rhythm / T-helper 1 cell lineage commitment / Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S / positive regulation of pentose-phosphate shunt / positive regulation of wound healing, spreading of epidermal cells / eukaryotic initiation factor 4E binding ...regulation of type B pancreatic cell development / RNA polymerase III type 2 promoter sequence-specific DNA binding / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / regulation of locomotor rhythm / T-helper 1 cell lineage commitment / Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S / positive regulation of pentose-phosphate shunt / positive regulation of wound healing, spreading of epidermal cells / eukaryotic initiation factor 4E binding / TORC2 complex / regulation of membrane permeability / TORC2 signaling / cellular response to leucine starvation / TFIIIC-class transcription factor complex binding / heart valve morphogenesis / negative regulation of lysosome organization / TORC1 complex / voluntary musculoskeletal movement / positive regulation of transcription of nucleolar large rRNA by RNA polymerase I / positive regulation of odontoblast differentiation / calcineurin-NFAT signaling cascade / RNA polymerase III type 3 promoter sequence-specific DNA binding / positive regulation of keratinocyte migration / regulation of osteoclast differentiation / MTOR signalling / regulation of lysosome organization / cellular response to L-leucine / energy reserve metabolic process / regulation of autophagosome assembly / Energy dependent regulation of mTOR by LKB1-AMPK / cellular response to nutrient / Amino acids regulate mTORC1 / cellular response to methionine / serine/threonine protein kinase complex / ruffle organization / negative regulation of cell size / positive regulation of ubiquitin-dependent protein catabolic process / positive regulation of osteoclast differentiation / cellular response to osmotic stress / anoikis / negative regulation of cold-induced thermogenesis / inositol hexakisphosphate binding / negative regulation of protein localization to nucleus / cardiac muscle cell development / negative regulation of calcineurin-NFAT signaling cascade / regulation of myelination / positive regulation of transcription by RNA polymerase III / small GTPase-mediated signal transduction / negative regulation of macroautophagy / TORC1 signaling / positive regulation of myotube differentiation / Macroautophagy / regulation of cell size / Constitutive Signaling by AKT1 E17K in Cancer / positive regulation of actin filament polymerization / germ cell development / behavioral response to pain / oligodendrocyte differentiation / positive regulation of oligodendrocyte differentiation / TOR signaling / positive regulation of translational initiation / mTORC1-mediated signalling / CD28 dependent PI3K/Akt signaling / HSF1-dependent transactivation / regulation of macroautophagy / social behavior / positive regulation of TOR signaling / enzyme-substrate adaptor activity / positive regulation of G1/S transition of mitotic cell cycle / protein serine/threonine kinase inhibitor activity / protein kinase activator activity / 'de novo' pyrimidine nucleobase biosynthetic process / response to amino acid / positive regulation of epithelial to mesenchymal transition / positive regulation of lipid biosynthetic process / vascular endothelial cell response to laminar fluid shear stress / heart morphogenesis / neuronal action potential / regulation of cellular response to heat / positive regulation of lamellipodium assembly / cardiac muscle contraction / positive regulation of stress fiber assembly / T cell costimulation / phagocytic vesicle / 14-3-3 protein binding / positive regulation of endothelial cell proliferation / translation repressor activity / translation initiation factor binding / negative regulation of translational initiation / cellular response to nutrient levels / cytoskeleton organization / endomembrane system / positive regulation of TORC1 signaling / negative regulation of insulin receptor signaling pathway / positive regulation of mitotic cell cycle / negative regulation of autophagy / cellular response to amino acid starvation / cellular response to starvation / positive regulation of glycolytic process Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Pavletich NP / Yang H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Authors: Haijuan Yang / Xiaolu Jiang / Buren Li / Hyo J Yang / Meredith Miller / Angela Yang / Ankita Dhar / Nikola P Pavletich /  Abstract: The mechanistic target of rapamycin complex 1 (mTORC1) controls cell growth and metabolism in response to nutrients, energy levels, and growth factors. It contains the atypical kinase mTOR and the ...The mechanistic target of rapamycin complex 1 (mTORC1) controls cell growth and metabolism in response to nutrients, energy levels, and growth factors. It contains the atypical kinase mTOR and the RAPTOR subunit that binds to the Tor signalling sequence (TOS) motif of substrates and regulators. mTORC1 is activated by the small GTPase RHEB (Ras homologue enriched in brain) and inhibited by PRAS40. Here we present the 3.0 ångström cryo-electron microscopy structure of mTORC1 and the 3.4 ångström structure of activated RHEB-mTORC1. RHEB binds to mTOR distally from the kinase active site, yet causes a global conformational change that allosterically realigns active-site residues, accelerating catalysis. Cancer-associated hyperactivating mutations map to structural elements that maintain the inactive state, and we provide biochemical evidence that they mimic RHEB relieving auto-inhibition. We also present crystal structures of RAPTOR-TOS motif complexes that define the determinants of TOS recognition, of an mTOR FKBP12-rapamycin-binding (FRB) domain-substrate complex that establishes a second substrate-recruitment mechanism, and of a truncated mTOR-PRAS40 complex that reveals PRAS40 inhibits both substrate-recruitment sites. These findings help explain how mTORC1 selects its substrates, how its kinase activity is controlled, and how it is activated by cancer-associated mutations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7086.map.gz emd_7086.map.gz | 186.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7086-v30.xml emd-7086-v30.xml emd-7086.xml emd-7086.xml | 26 KB 26 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7086.png emd_7086.png | 62.2 KB | ||
| Filedesc metadata |  emd-7086.cif.gz emd-7086.cif.gz | 8.8 KB | ||
| Others |  emd_7086_additional_1.map.gz emd_7086_additional_1.map.gz emd_7086_additional_2.map.gz emd_7086_additional_2.map.gz emd_7086_additional_3.map.gz emd_7086_additional_3.map.gz | 186.8 MB 186.8 MB 186.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7086 http://ftp.pdbj.org/pub/emdb/structures/EMD-7086 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7086 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7086 | HTTPS FTP |
-Related structure data
| Related structure data |  6bcuMC  7087C  5wbhC  5wbiC  5wbjC  5wbkC  5wblC  5wbuC  5wbyC  6bcxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7086.map.gz / Format: CCP4 / Size: 199.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7086.map.gz / Format: CCP4 / Size: 199.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary consensus dimer map in C2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3306 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Focused 3D refinement of monomer part2 (mask 2)...
| File | emd_7086_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused 3D refinement of monomer part2 (mask 2) used to reconstruct the 3.4 angstroms resolution C2 dimer structure factors with the REFMAC5 composite map option for model refinement. | ||||||||||||
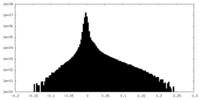
| Projections & Slices |
| ||||||||||||
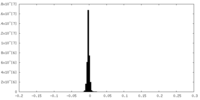
| Density Histograms |
-Additional map: Focused 3D refinement of monomer part1 (mask 1)...
| File | emd_7086_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused 3D refinement of monomer part1 (mask 1) used to reconstruct the 3.4 angstroms resolution C2 dimer structure factors with the REFMAC5 composite map option for model refinement. | ||||||||||||
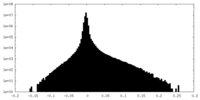
| Projections & Slices |
| ||||||||||||
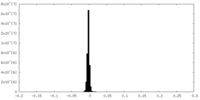
| Density Histograms |
-Additional map: Focused 3D refinement of monomer part3 (mask 3)...
| File | emd_7086_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused 3D refinement of monomer part3 (mask 3) used to reconstruct the 3.4 angstroms resolution C2 dimer structure factors with the REFMAC5 composite map option for model refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : active complex
| Entire | Name: active complex |
|---|---|
| Components |
|
-Supramolecule #1: active complex
| Supramolecule | Name: active complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Serine/threonine-protein kinase mTOR,Serine/threonine-protein kin...
| Macromolecule | Name: Serine/threonine-protein kinase mTOR,Serine/threonine-protein kinase mTOR type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 287.399125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLGTGPAAAT TAATTS(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)SRNEET(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)EMSQE ...String: MLGTGPAAAT TAATTS(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)SRNEET(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)EMSQE E(UNK)TRFYDQLN HHIFELVSSS DANERKGGIL AIASLIGVEG GNATRI GRF ANYLRNLLPS NDPVVMEMAS KAIGRLAMAG DTFTAEYVEF EVKRALEWLG ADRNEGRRHA AVLVLRELAI SVPTFFF QQ VQPFFDNIFV AVWDPKQAIR EGAVAALRAC LILTTQREPK EMQKPQWYRH TFEEAEKGFD ETLAKEKGMN RDDRIHGA L LILNELVRIS SMEGERLREE MEEITQQQLV HDKYCKDLMG FGTKPRHITP FTSFQAVQPQ QSNALVGLLG YSSHQGLMG FGTSPSPAKS T(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)RNS KNSLIQMTIL NLLPRLA AF RPSAFTDTQY LQDTMNHVLS CVKKEKERTA AFQALGLLSV AVRSEFKVYL PRVLDIIRAA LPPKDFAHKR QKAMQVDA T VFTCISMLAR AMGPGIQQDI KELLEPMLAV GLSPALTAVL YDLSRQIPQL KKDIQDGLLK MLSLVLM(UNK)KP LRHPG MPKG LAHQLASPGL TTLPEAS(UNK)VG SITLALRTLG SFEFEGHSLT QFVRHCADHF LNSEHKEIRM EAARTCSRLL TP SIHLISG HAHVVSQTAV QVVADVLSKL LVVGITDPDP DIRYCVLASL DERFDAHLAQ AENLQALFVA LNDQVFEIRE LAI CTVGRL SSMNPAFVMP FLRKMLIQIL TELEHSGIGR IKEQSARMLG HLVSNAPRLI RPYMEPILKA LILKLKDPDP DPNP GVINN VLATIGELAQ VSGLEMRKWV DELFIIIMDM LQDSSLLAKR QVALWTLGQL VASTGYVVEP YRKYPTLLEV LLNFL KTEQ NQGTRREAIR VLGLLGALDP YKHKVNIGMI DQSRDASAVS LSESKSSQDS SDYSTSEMLV NMGNLPLDEF YPAVSM VAL MRIFRDQSLS HHHTMVVQAI TFIFKSLGLK CVQFLPQVMP TFLNVIRVCD GAIREFLFQQ LGMLVSFVKS HIRPYMD EI VTLMREFWVM NTSIQSTIIL LIEQIVVALG GEFKLYLPQL IPHMLRVFMH DNSPGRIVSI KLLAAIQLFG ANLDDYLH L LLPPIVKLFD APEAPLPSRK AALETVDRLT ESLDFTDYAS RIIHPIVRTL DQSPELRSTA MDTLSSLVFQ LGKKYQIFI PMVNKVLVRH RINHQRYDVL ICRIVKGYTL ADEEEDPLIY QHRMLRSGQG DALASGPVET GPMKKLHVST INLQKAWGAA RRVSKDDWL EWLRRLSLEL LKDSSSPSLR SCWALAQAYN PMARDLFNAA FVSCWSELNE DQQDELIRSI ELALTSQDIA E VTQTLLNL AEFMEHSDKG PLPLRDDNGI VLLGERAAKC RAYAKALHYK ELEFQKGPTP AILESLISIN NKLQQPEAAA GV LEYAMKH FGELEIQATW YEKLHEWEDA LVAYDKKMDT NKDDPELMLG RMRCLEALGE WGQLHQQCCE KWTLVNDETQ AKM ARMAAA AAWGLGQWDS MEEYTCMIPR DTHDGAFYRA VLALHQDLFS LAQQCIDKAR DLLDAELTAM AGESYSRAYG AMVS CHMLS ELEEVIQYKL VPERREIIRQ IWWERLQGCQ RIVEDWQKIL MVRSLVVSPH EDMRTWLKYA SLCGKSGRLA LAHKT LVLL LGVDPSRQLD HPLPTVHPQV TYAYMKNMWK SARKIDAFQH MQHFVQTMQQ QAQHAIATED QQHKQELHKL MARCFL KLG EWQLNLQGIN ESTIPKVLQY YSAATEHDRS WYKAWHAWAV MNFEAVLHYK HQNQARDEKK KLRHASGANI TNATTAA TT AATATTTAST EGSNSESEAE STENSPTPSP LQKKVTEDLS KTLLMYTVPA VQGFFRSISL SRGNNLQDTL RVLTLWFD Y GHWPDVNEAL VEGVKAIQID TWLQVIPQLI ARIDTPRPLV GRLIHQLLTD IGRYHPQALI YPLTVASKST TTARHNAAN KILKNMCEHS NTLVQQAMMV SEELIRVAIL WHEMWHEGLE EASRLYFGER NVKGMFEVLE PLHAMMERGP QTLKETSFNQ AYGRDLMEA QEWCRKYMKS GNVKDLTQAW DLYYHVFRRI SKQLPQLTSL ELQYVSPKLL MCRDLELAVP GTYDPNQPII R IQSIAPSL QVITSKQRPR KLTLMGSNGH EFVFLLKGHE DLRQDERVMQ LFGLVNTLLA NDPTSLRKNL SIQRYAVIPL ST NSGLIGW VPHCDTLHAL IRDYREKKKI LLNIEHRIML RMAPDYDHLT LMQKVEVFEH AVNNTAGDDL AKLLWLKSPS SEV WFDRRT NYTRSLAVMS MVGYILGLGD RHPSNLMLDR LSGKILHIDF GDCFEVAMTR EKFPEKIPFR LTRMLTNAME VTGL DGNYR ITCHTVMEVL REHKDSVMAV LEAFVYDPLL NWRLMDTNTK GNKRSRTRTD SYSAGQSVEI LDGVELGEPA HKKTG TTVP ESIHSFIGDG LVKPEALNKK AIQIINRVRD KLTGRDFSHD DTLDVPTQVE LLIKQATSHE NLCQCYIGWC PFW UniProtKB: Serine/threonine-protein kinase mTOR |
-Macromolecule #2: Target of rapamycin complex subunit LST8
| Macromolecule | Name: Target of rapamycin complex subunit LST8 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.91009 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE ...String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE PEVSITSAHI DPDASYMAAV NSTGNCYVWN LTGGIGDEVT QLIPKTKIPA HTRYALQCRF SPDSTLLATC SA DQTCKIW RTSNFSLMTE LSIKSGNPGE SSRGWMWGCA FSGDSQYIVT ASSDNLARLW CVETGEIKRE YGGHQKAVVC LAF NDSVLG UniProtKB: Target of rapamycin complex subunit LST8 |
-Macromolecule #3: Regulatory-associated protein of mTOR,Regulatory-associated prote...
| Macromolecule | Name: Regulatory-associated protein of mTOR,Regulatory-associated protein of mTOR type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150.197 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKE SEMLQSPLLG LGEEDEADLT DWNLPLAFMK KRHCEKIEGS KSLAQSWRMK DRMKTVSVAL VLCLNVGVDP PDVVKTTPC ARLECWIDPL SMGPQKALET IGANLQKQYE NWQPRARYKQ SLDPTVDEVK KLCTSLRRNA KEERVLFHYN G HGVPRPTV ...String: MDYKDDDDKE SEMLQSPLLG LGEEDEADLT DWNLPLAFMK KRHCEKIEGS KSLAQSWRMK DRMKTVSVAL VLCLNVGVDP PDVVKTTPC ARLECWIDPL SMGPQKALET IGANLQKQYE NWQPRARYKQ SLDPTVDEVK KLCTSLRRNA KEERVLFHYN G HGVPRPTV NGEVWVFNKN YTQYIPLSIY DLQTWMGSPS IFVYDCSNAG LIVKSFKQFA LQREQELEVA AINPNHPLAQ MP LPPSMKN CIQLAACEAT ELLPMIPDLP ADLFTSCLTT PIKIALRWFC MQKCVSLVPG VTLDLIEKIP GRLNDRRTPL GEL NWIFTA ITDTIAWNVL PRDLFQKLFR QDLLVASLFR NFLLAERIMR SYNCTPVSSP RLPPTYMHAM WQAWDLAVDI CLSQ LPTII EEGTAFRHSP FFAEQLTAFQ VWLTMGVENR NPPEQLPIVL QVLLSQVHRL RALDLLGRFL DLGPWAVSLA LSVGI FPYV LKLLQSSARE LRPLLVFIWA KILAVDSSCQ ADLVKDNGHK YFLSVLADPY MPAEHRTMTA FILAVIVNSY HTGQEA CLQ GNLIAICLEQ LNDPHPLLRQ WVAICLGRIW QNFDSARWCG VRDSAHEKLY SLLSDPIPEV RCAAVFALGT FVGNSAE RT DHSTTIDHNV AMMLAQLVSD GSPMVRKELV VALSHLVVQY ESNFCTVALQ FIEEEKNYAL PSPATTEGGS LTPVRDSP C TPRLRSVSSY GNIRAVATAR SLNKSLQNLS LTEESGGAVA FSPGNLSTSS SASSTLGSPE NEEHILSFET IDKMRRASS YSSLNSLIGV SFNSVYTQIW RVLLHLAADP YPEVSDVAMK VLNSIAYKAT VNARPQRVLD TSSLTQSAPA SPTNKGVHIH QAGGSPPAS STSSSSLTND VAKQPVSRDL PSGRPGTTGP AGAQYTPHSH QFPRTRKMFD KGPEQTADDA DDAAGHKSFI S ATVQTGFC DWSARYFAQP VMKIPEEHDL ESQIRKEREW RFLRNSRVRR QAQQVIQKGI TRLDDQIFLN RNPGVPSVVK FH PFTPCIA VADKDSICFW DWEKGEKLDY FHNGNPRYTR VTAMEYLNGQ DCSLLLTATD DGAIRVWKNF ADLEKNPEMV TAW QGLSDM LPTTRGAGMV VDWEQETGLL MSSGDVRIVR IWDTDREMKV QDIPTGADSC VTSLSCDSHR SLIVAGLGDG SIRV YDRRM ALSECRVMTY REHTAWVVKA SLQKRPDGHI VSVSVNGDVR IFDPRMPESV NVLQIVKGLT ALDIHPQADL IACGS VNQF TAIYNSSGEL INNIKYYDGF MGQRVGAISC LAFHPHWPHL AVGSNDYYIS VYSVEKRVR UniProtKB: Regulatory-associated protein of mTOR, Regulatory-associated protein of mTOR |
-Macromolecule #4: Eukaryotic translation initiation factor 4E-binding protein 1
| Macromolecule | Name: Eukaryotic translation initiation factor 4E-binding protein 1 type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.951344 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSGRMSGGSS CSQTPSRAIP ATRRVVLGDG VQLPPGDYST TPGGTLFSTT PGGTRIIYDR KFLMECRNSP VTKTPPRDLP TIPGVTSPS SDEPPMEASQ SHLRNSPEDK RAGGEESQFE MDI UniProtKB: Eukaryotic translation initiation factor 4E-binding protein 1 |
-Macromolecule #5: GTP-binding protein Rheb
| Macromolecule | Name: GTP-binding protein Rheb / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.877826 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSGRMPQSKS RKIAILGYRS VGKSSLTIQF VEGQFVDSYD PTIENTFTKL ITVNGQEYHL QLVDTAGQDE YSIFPQTYSI DINGYILVY SVTSIKSFEV IKVIHGKLLD MVGKVQIPIM LVGNKKDLHM ERVISYEEGK ALAESWNAAF LESSAKENQT A VDVFRRII LEAEKMDGAA SQGKSSCSVM UniProtKB: GTP-binding protein Rheb |
-Macromolecule #6: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #8: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 2 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2) / Number images used: 198237 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6bcu: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)