+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7055 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the NAIP5-NLRC4-flagellin inflammasome | ||||||||||||
 Map data Map data | Postprocessed (sharpened, filtered) final map of the NAIP5-NLRC4-flagellin inflammasome | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Innate immunity / molecular complex / IMMUNE SYSTEM | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationIPAF inflammasome complex / caspase binding / positive regulation of protein processing / bacterial-type flagellum / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / pyroptotic inflammatory response / endopeptidase activator activity / detection of bacterium / activation of innate immune response / positive regulation of interleukin-1 beta production ...IPAF inflammasome complex / caspase binding / positive regulation of protein processing / bacterial-type flagellum / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / pyroptotic inflammatory response / endopeptidase activator activity / detection of bacterium / activation of innate immune response / positive regulation of interleukin-1 beta production / protein homooligomerization / regulation of apoptotic process / defense response to Gram-negative bacterium / defense response to bacterium / positive regulation of apoptotic process / inflammatory response / innate immune response / intracellular membrane-bounded organelle / apoptotic process / symbiont entry into host cell / negative regulation of apoptotic process / structural molecule activity / protein homodimerization activity / extracellular region / ATP binding / metal ion binding / identical protein binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | ||||||||||||
 Authors Authors | Tenthorey JL / Haloupek N | ||||||||||||
| Funding support |  Spain, Spain,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Authors: Jeannette L Tenthorey / Nicole Haloupek / José Ramón López-Blanco / Patricia Grob / Elise Adamson / Ella Hartenian / Nicholas A Lind / Natasha M Bourgeois / Pablo Chacón / Eva Nogales / Russell E Vance /   Abstract: Robust innate immune detection of rapidly evolving pathogens is critical for host defense. Nucleotide-binding domain leucine-rich repeat (NLR) proteins function as cytosolic innate immune sensors in ...Robust innate immune detection of rapidly evolving pathogens is critical for host defense. Nucleotide-binding domain leucine-rich repeat (NLR) proteins function as cytosolic innate immune sensors in plants and animals. However, the structural basis for ligand-induced NLR activation has so far remained unknown. NAIP5 (NLR family, apoptosis inhibitory protein 5) binds the bacterial protein flagellin and assembles with NLRC4 to form a multiprotein complex called an inflammasome. Here we report the cryo-electron microscopy structure of the assembled ~1.4-megadalton flagellin-NAIP5-NLRC4 inflammasome, revealing how a ligand activates an NLR. Six distinct NAIP5 domains contact multiple conserved regions of flagellin, prying NAIP5 into an open and active conformation. We show that innate immune recognition of multiple ligand surfaces is a generalizable strategy that limits pathogen evolution and immune escape. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7055.map.gz emd_7055.map.gz | 321 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7055-v30.xml emd-7055-v30.xml emd-7055.xml emd-7055.xml | 25.6 KB 25.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7055.png emd_7055.png | 50.8 KB | ||
| Masks |  emd_7055_msk_1.map emd_7055_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-7055.cif.gz emd-7055.cif.gz | 8.4 KB | ||
| Others |  emd_7055_half_map_1.map.gz emd_7055_half_map_1.map.gz emd_7055_half_map_2.map.gz emd_7055_half_map_2.map.gz | 273.6 MB 273.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7055 http://ftp.pdbj.org/pub/emdb/structures/EMD-7055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7055 | HTTPS FTP |
-Validation report
| Summary document |  emd_7055_validation.pdf.gz emd_7055_validation.pdf.gz | 910.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7055_full_validation.pdf.gz emd_7055_full_validation.pdf.gz | 910.1 KB | Display | |
| Data in XML |  emd_7055_validation.xml.gz emd_7055_validation.xml.gz | 17.1 KB | Display | |
| Data in CIF |  emd_7055_validation.cif.gz emd_7055_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7055 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7055 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7055 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7055 | HTTPS FTP |
-Related structure data
| Related structure data |  6b5bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7055.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7055.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed (sharpened, filtered) final map of the NAIP5-NLRC4-flagellin inflammasome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
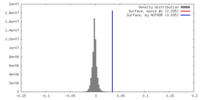
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7055_msk_1.map emd_7055_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
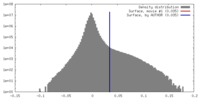
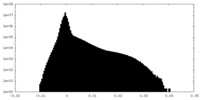
| Density Histograms |
-Half map: Unfiltered, unsharpened half map #1 of the NAIP5-NLRC4-flagellin...
| File | emd_7055_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered, unsharpened half map #1 of the NAIP5-NLRC4-flagellin inflammasome | ||||||||||||
| Projections & Slices |
| ||||||||||||
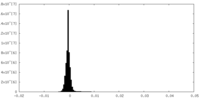
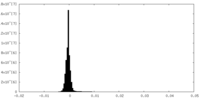
| Density Histograms |
-Half map: Unfiltered, unsharpened half map #2 of the NAIP5-NLRC4-flagellin...
| File | emd_7055_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered, unsharpened half map #2 of the NAIP5-NLRC4-flagellin inflammasome | ||||||||||||
| Projections & Slices |
| ||||||||||||
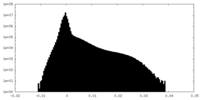
| Density Histograms |
- Sample components
Sample components
-Entire : NAIP5-NLRC4-flagellin inflammasome
| Entire | Name: NAIP5-NLRC4-flagellin inflammasome |
|---|---|
| Components |
|
-Supramolecule #1: NAIP5-NLRC4-flagellin inflammasome
| Supramolecule | Name: NAIP5-NLRC4-flagellin inflammasome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: NAIP5
| Supramolecule | Name: NAIP5 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: NLRC4
| Supramolecule | Name: NLRC4 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: flagellin
| Supramolecule | Name: flagellin / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Baculoviral IAP repeat-containing protein 1e
| Macromolecule | Name: Baculoviral IAP repeat-containing protein 1e / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 159.874828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEHGESSED RISEIDYEFL PELSALLGVD AFQVAKSQEE EEHKERMKMK KGFNSQMRSE AKRLKTFETY DTFRSWTPQE MAAAGFYHT GVKLGVQCFC CSLILFGNSL RKLPIERHKK LRPECEFLQG KDVGNIGKYD IRVKSPEKML RGGKARYHEE E ARLESFED ...String: MAEHGESSED RISEIDYEFL PELSALLGVD AFQVAKSQEE EEHKERMKMK KGFNSQMRSE AKRLKTFETY DTFRSWTPQE MAAAGFYHT GVKLGVQCFC CSLILFGNSL RKLPIERHKK LRPECEFLQG KDVGNIGKYD IRVKSPEKML RGGKARYHEE E ARLESFED WPFYAHGTSP RVLSAAGFVF TGKRDTVQCF SCGGSLGNWE EGDDPWKEHA KWFPKCEFLQ SKKSSEEIAQ YI QSYEGFV HVTGEHFVKS WVRRELPMVS AYCNDSVFAN EELRMDMFKD WPQESPVGVE ALVRAGFFYT GKKDIVRCFS CGG CLEKWA EGDDPMEDHI KFFPECVFLQ TLKSSAEVIP TLQSQYALPE ATETTRESNH GDAAAVHSTV VDLGRSEAQW FQEA RSLSE QLRDNYTKAT FRHMNLPEVC SSLGTDHLLS CDVSIISKHI SQPVQEALTI PEVFSNLNSV MCVEGETGSG KTTFL KRIA FLWASGCCPL LYRFQLVFYL SLSSITPDQG LANIICAQLL GAGGCISEVC LSSSIQQLQH QVLFLLDDYS GLASLP QAL HTLITKNYLS RTCLLIAVHT NRVRDIRLYL GTSLEIQEFP FYNTVSVLRK FFSHDIICVE KLIIYFIDNK DLQGVYK TP LFVAAVCTDW IQNASAQDKF QDVTLFQSYM QYLSLKYKAT AEPLQATVSS CGQLALTGLF SSCFEFNSDD LAEAGVDE D EKLTTLLMSK FTAQRLRPVY RFLGPLFQEF LAAVRLTELL SSDRQEDQDL GLYYLRQIDS PLKAINSFNI FLYYVSSHS SSKAAPTVVS HLLQLVDEKE SLENMSENED YMKLHPQTFL WFQFVRGLWL VSPESSSSFV SEHLLRLALI FAYESNTVAE CSPFILQFL RGKTLALRVL NLQYFRDHPE SLLLLRSLKV SINGNKMSSY VDYSFKTYFE NLQPPAIDEE YTSAFEHISE W RRNFAQDE EIIKNYENIR PRALPDISEG YWKLSPKPCK IPKLEVQVNN TDAADQALLQ VLMEVFSASQ SIEFRLFNSS GF LESICPA LELSKASVTK CSMSRLELSR AEQELLLTLP ALQSLEVSET NQLPEQLFHN LHKFLGLKEL CVRLDGKPDV LSV LPGEFP NLHHMEKLSI RTSTESDLSK LVKFIQNFPN LHVFHLKCDF LSNCESLMAV LASCKKLREI EFSGRCFEAM TFVN ILPNF VSLKILNLKD QQFPDKETSE KFAQALGSLR NLEELLVPTG DGIHQVAKLI VRQCLQLPCL RVLTFHDILD DDSVI EIAR AATSGGFQKL ENLDISMNHK ITEEGYRNFF QALDNLPNLQ ELNICRNIPG RIQVQATTVK ALGQCVSRLP SLIRLH MLS WLLDEEDMKV INDVKERHPQ SKRLIIFWKL IVPFSPVILE UniProtKB: Baculoviral IAP repeat-containing protein 1e |
-Macromolecule #2: NLR family CARD domain-containing protein 4
| Macromolecule | Name: NLR family CARD domain-containing protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 116.886 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNFIRNNRRA LIQRMGLTVT KQICDDLFAL NVLNNQEANV IYCEPLEQEA ARKIIHMTMQ KGSAACNLFL KSLENWDYFV YQDLTGQNL SYQVTEEDLN VLAQNLKDLY NSPAFLNFYP LGEDIDIIFN LEKTFTEPIM WKKDHRHHRV EQLTLGSLLE A LKSPCLIE ...String: MNFIRNNRRA LIQRMGLTVT KQICDDLFAL NVLNNQEANV IYCEPLEQEA ARKIIHMTMQ KGSAACNLFL KSLENWDYFV YQDLTGQNL SYQVTEEDLN VLAQNLKDLY NSPAFLNFYP LGEDIDIIFN LEKTFTEPIM WKKDHRHHRV EQLTLGSLLE A LKSPCLIE GESGKGKSTL LQRIAMLWAS GGCRALKGFR LVFFIHLRSA RGGLFETLYD QLLNIPDFIS KPTFKALLLK LH KEVLFLL DGYNEFHPQN CPEIEALIKE NHRFKNMVIV TTTTECLRHI RHVGALTAEV GDMTEDSAKD LIEAVLVPDQ VER LWAQIQ ESRCLRNLMK TPLFVVITCA IQMGRQEFQA HTQTMLFQTF YDLLIQKNSH RYRGGASGDF ARSLDYCGDL ALEG VFAHK FDFEPEHGSS MNEDVLVTIG LLCKYTAQRL KPTYKFFHKS FQEYTAGRRL SSLLTSKEPE EVSKGNSYLN KMVSI SDIT SLYGNLLLYT CGSSTEATRA VMRHLAMVYQ HGSLQGLSVT KRPLWRQESI QSLRNTTEQD VLKAINVNSF VECGIN LFS ESMSKSDLSQ EFEAFFQGKS LYINSENIPD YLFDFFEYLP NCASALDFVK LDFYERATES QDKAEENVPG VHTEGPS ET YIPPRAVSLF FNWKQEFKTL EVTLRDINKL NKQDIKYLGK IFSSATNLRL HIKRCAAMAG RLSSVLRTCK NMHTLMVE A SPLTTDDEQY ITSVTGLQNL SIHRLHTQQL PGGLIDSLGN LKNLERLILD DIRMNEEDAK NLAEGLRSLK KMRLLHLTH LSDIGEGMDY IVKSLSEESC DLQEMKLVAC CLTANSVKVL AQNLHNLIKL SILDISENYL EKDGNEALQE LIGRLGVLGE LTTLMLPWC WDVHTSLPKL LKQLEGTPGL AKLGLKNWRL RDEEIKSLGE FLEMNPLRDL QQLDLAGHCV SSDGWLYFMN V FENLKQLV FFDFSTEEFL PDAALVRKLS QVLSKLTLLQ EVKLTGWEFD DYDISAIKGT FKLVTA UniProtKB: NLR family CARD domain-containing protein 4 |
-Macromolecule #3: Flagellin
| Macromolecule | Name: Flagellin / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 58.49407 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEQKLISEED LNEMEQKLIS EEDLNEMEQK LISEEDLNEM EQKLISEEDL NEMEQKLISE EDLNEMESLG DLTMEQKLIS EEDLNSGRP AAMAQVINTN VASLTAQRNL GVSGNMMQTS IQRLSSGLRI NSAKDDAAGL AISQRMTAQI RGMNQAVRNA N DGISLAQV ...String: MEQKLISEED LNEMEQKLIS EEDLNEMEQK LISEEDLNEM EQKLISEEDL NEMEQKLISE EDLNEMESLG DLTMEQKLIS EEDLNSGRP AAMAQVINTN VASLTAQRNL GVSGNMMQTS IQRLSSGLRI NSAKDDAAGL AISQRMTAQI RGMNQAVRNA N DGISLAQV AEGAMQETTN ILQRMRELSV QAANSTNNSS DRSSIQSEIS QLKSELERIA QNTEFNGQRI LDGSFSGASF QV GANSNQT INFSIGSTKA SSLGGIATAT GTEVAGAAAA DITIAIGGGA ATSINSSANF TGALNGQDAT SAYAKAAAIN DAG IGGLSV TASTSGTQAV GAIGGTAGDT YNLTINGVAI YTNLNVATAL TNSDLRDAIN GVSNQTGVVA SLNGGNMTLT AADG RNITV TESGTGFTAG TDGLTVTGGA FDGALRGTLS ISAVDTIAIG GTVANIGLSA NISKDTVGID SLDVSTASGA QTAIK RIDA ALNSVNSNRA NMGALQNRFE STIANLQNVS DNLSAARSRI QDADYAAEMA SLTKNQILQQ AGTAMLAQAN SLPQSV LSL LGR UniProtKB: UNIPROTKB: G8UUW9 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Component:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Details: Carbon-coated | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Frames/image: 1-20 / Number grids imaged: 3 / Average exposure time: 6.0 sec. / Average electron dose: 45.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 22500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Homology models predicted by I-TASSER server were used as initial model sources. The main structural template identified by I-TASSER for NAIP5 was the crystal structure of NLRC4 in the inactive conformation (PDB ID: 4KXF) that covered all the domains except the N-terminal BIR region, where homology models from several BIR domains were recognized (PDB IDs: 1SE0, 2VM5, and 1OXQ for BIR1, BIR2, and BIR3, respectively). Predictions were first rigid body docked into the density map using Chimera and/or ADP_EM. Then, the docked models were flexibly fitted with iMODFIT, if necessary. Finally, all models were refined in Phenix. |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 167 / Target criteria: Correlation coefficient |
| Output model |  PDB-6b5b: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)