[English] 日本語
 Yorodumi
Yorodumi- EMDB-7009: Structure of an acid sensing ion channel in a resting state at high pH -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7009 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of an acid sensing ion channel in a resting state at high pH | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Ion channel / ASIC / ASIC1a / Sodium channel / TRANSPORT PROTEIN / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationStimuli-sensing channels / pH-gated monoatomic ion channel activity / cellular response to pH / ligand-gated sodium channel activity / sodium ion transmembrane transport / postsynaptic membrane / dendrite / identical protein binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Yoder N / Yoshioka C | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Gating mechanisms of acid-sensing ion channels. Authors: Nate Yoder / Craig Yoshioka / Eric Gouaux /  Abstract: Acid-sensing ion channels (ASICs) are trimeric, proton-gated and sodium-selective members of the epithelial sodium channel/degenerin (ENaC/DEG) superfamily of ion channels and are expressed ...Acid-sensing ion channels (ASICs) are trimeric, proton-gated and sodium-selective members of the epithelial sodium channel/degenerin (ENaC/DEG) superfamily of ion channels and are expressed throughout vertebrate central and peripheral nervous systems. Gating of ASICs occurs on a millisecond time scale and the mechanism involves three conformational states: high pH resting, low pH open and low pH desensitized. Existing X-ray structures of ASIC1a describe the conformations of the open and desensitized states, but the structure of the high pH resting state and detailed mechanisms of the activation and desensitization of the channel have remained elusive. Here we present structures of the high pH resting state of homotrimeric chicken (Gallus gallus) ASIC1a, determined by X-ray crystallography and single particle cryo-electron microscopy, and present a comprehensive molecular mechanism for proton-dependent gating in ASICs. In the resting state, the position of the thumb domain is further from the three-fold molecular axis, thereby expanding the 'acidic pocket' in comparison to the open and desensitized states. Activation therefore involves 'closure' of the thumb into the acidic pocket, expansion of the lower palm domain and an iris-like opening of the channel gate. Furthermore, we demonstrate how the β11-β12 linkers that demarcate the upper and lower palm domains serve as a molecular 'clutch', and undergo a simple rearrangement to permit rapid desensitization. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7009.map.gz emd_7009.map.gz | 23.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7009-v30.xml emd-7009-v30.xml emd-7009.xml emd-7009.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7009.png emd_7009.png | 148.5 KB | ||
| Filedesc metadata |  emd-7009.cif.gz emd-7009.cif.gz | 6.1 KB | ||
| Others |  emd_7009_additional.map.gz emd_7009_additional.map.gz emd_7009_half_map_1.map.gz emd_7009_half_map_1.map.gz emd_7009_half_map_2.map.gz emd_7009_half_map_2.map.gz | 28.3 MB 13.8 MB 13.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7009 http://ftp.pdbj.org/pub/emdb/structures/EMD-7009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7009 | HTTPS FTP |
-Related structure data
| Related structure data |  6aveMC  5wkuC  5wkvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7009.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7009.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
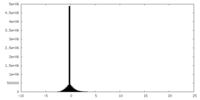
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_7009_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
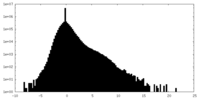
| Density Histograms |
-Half map: #1
| File | emd_7009_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
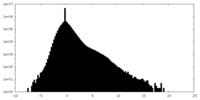
| Density Histograms |
-Half map: #2
| File | emd_7009_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Acid Sensing Ion Channel 1a
| Entire | Name: Acid Sensing Ion Channel 1a |
|---|---|
| Components |
|
-Supramolecule #1: Acid Sensing Ion Channel 1a
| Supramolecule | Name: Acid Sensing Ion Channel 1a / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 180.03144 KDa |
-Macromolecule #1: Acid-sensing ion channel 1
| Macromolecule | Name: Acid-sensing ion channel 1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.080324 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMDLKVDEEE VDSGQPVSIQ AFASSSTLHG ISHIFSYERL SLKRVVWALC FMGSLALLAL VCTNRIQYYF LYPHVTKLDE VAATRLTFP AVTFCNLNEF RFSRVTKNDL YHAGELLALL NNRYEIPDTQ TADEKQLEIL QDKANFRNFK PKPFNMLEFY D RAGHDIRE ...String: MMDLKVDEEE VDSGQPVSIQ AFASSSTLHG ISHIFSYERL SLKRVVWALC FMGSLALLAL VCTNRIQYYF LYPHVTKLDE VAATRLTFP AVTFCNLNEF RFSRVTKNDL YHAGELLALL NNRYEIPDTQ TADEKQLEIL QDKANFRNFK PKPFNMLEFY D RAGHDIRE MLLSCFFRGE QCSPEDFKVV FTRYGKCYTF NAGQDGKPRL ITMKGGTGNG LEIMLDIQQD EYLPVWGETD ET SFEAGIK VQIHSQDEPP LIDQLGFGVA PGFQTFVSCQ EQRLIYLPPP WGDCKATTGD SEFYDTYSIT ACRIDCETRY LVE NCNCRM VHMPGDAPYC TPEQYKECAD PALDFLVEKD NEYCVCEMPC NVTRYGKELS MVKIPSKASA KYLAKKYNKS EQYI GENIL VLDIFFEALN YETIEQKKAY EVAGLLGDIG GQMGLFIGAS ILTVLELFDY AYEVIKHRLC RRGKCRKNHK RNNTD KGVA LSMDDVKRHN PCESLRGHPA GMTYAANILP HHPARGTFED FTC UniProtKB: Acid-sensing ion channel 1 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.2 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: 15 mA | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 26117 |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 2.1b1) |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: cisTEM |
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6ave: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)