+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6987 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a P-type ATPase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcalcium ion export across plasma membrane / regulation of receptor localization to synapse / calcium ion transmembrane transporter activity / type 1 fibroblast growth factor receptor binding / regulation of vascular associated smooth muscle contraction / excitatory synapse assembly / dendrite self-avoidance / cell-cell adhesion mediator activity / positive regulation of fibroblast growth factor receptor signaling pathway / positive regulation of long-term neuronal synaptic plasticity ...calcium ion export across plasma membrane / regulation of receptor localization to synapse / calcium ion transmembrane transporter activity / type 1 fibroblast growth factor receptor binding / regulation of vascular associated smooth muscle contraction / excitatory synapse assembly / dendrite self-avoidance / cell-cell adhesion mediator activity / positive regulation of fibroblast growth factor receptor signaling pathway / positive regulation of long-term neuronal synaptic plasticity / photoreceptor ribbon synapse / P-type Ca2+ transporter / P-type calcium transporter activity / GABA receptor activation / positive regulation of calcium ion transport / Sensory processing of sound by inner hair cells of the cochlea / Reduction of cytosolic Ca++ levels / negative regulation of cytokine production / negative regulation of cytosolic calcium ion concentration / molecular function inhibitor activity / homophilic cell adhesion via plasma membrane adhesion molecules / Ion transport by P-type ATPases / immunological synapse / regulation of cardiac conduction / regulation of cytosolic calcium ion concentration / positive regulation of bone mineralization / Ion homeostasis / cell adhesion molecule binding / regulation of cellular response to insulin stimulus / axon guidance / cell projection / positive regulation of neuron projection development / regulation of blood pressure / long-term synaptic potentiation / intracellular calcium ion homeostasis / synaptic vesicle membrane / positive regulation of protein phosphorylation / presynaptic membrane / positive regulation of cytosolic calcium ion concentration / monoatomic ion transmembrane transport / basolateral plasma membrane / calmodulin binding / postsynaptic density / axon / intracellular membrane-bounded organelle / glutamatergic synapse / cell surface / ATP hydrolysis activity / extracellular exosome / nucleoplasm / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
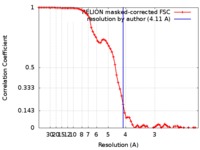
| Method | single particle reconstruction / cryo EM / Resolution: 4.11 Å | |||||||||
 Authors Authors | Gong DS / Chi XM | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structure of the human plasma membrane Ca-ATPase 1 in complex with its obligatory subunit neuroplastin. Authors: Deshun Gong / Ximin Chi / Kang Ren / Gaoxingyu Huang / Gewei Zhou / Nieng Yan / Jianlin Lei / Qiang Zhou /   Abstract: Plasma membrane Ca-ATPases (PMCAs) are key regulators of global Ca homeostasis and local intracellular Ca dynamics. Recently, Neuroplastin (NPTN) and basigin were identified as previously ...Plasma membrane Ca-ATPases (PMCAs) are key regulators of global Ca homeostasis and local intracellular Ca dynamics. Recently, Neuroplastin (NPTN) and basigin were identified as previously unrecognized obligatory subunits of PMCAs that dramatically increase the efficiency of PMCA-mediated Ca clearance. Here, we report the cryo-EM structure of human PMCA1 (hPMCA1) in complex with NPTN at a resolution of 4.1 Å for the overall structure and 3.9 Å for the transmembrane domain. The single transmembrane helix of NPTN interacts with the TM-linker and TM10 of hPMCA1. The subunits are required for the hPMCA1 functional activity. The NPTN-bound hPMCA1 closely resembles the E1-Mg structure of endo(sarco)plasmic reticulum Ca ATPase and the Ca site is exposed through a large open cytoplasmic pathway. This structure provides insight into how the subunits bind to the PMCAs and serves as an important basis for understanding the functional mechanisms of this essential calcium pump family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6987.map.gz emd_6987.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6987-v30.xml emd-6987-v30.xml emd-6987.xml emd-6987.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6987_fsc.xml emd_6987_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_6987.png emd_6987.png | 47.3 KB | ||
| Filedesc metadata |  emd-6987.cif.gz emd-6987.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6987 http://ftp.pdbj.org/pub/emdb/structures/EMD-6987 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6987 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6987 | HTTPS FTP |
-Related structure data
| Related structure data |  6a69MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6987.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6987.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
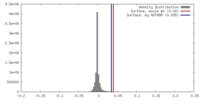
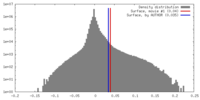
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : complex of one PMCA1 molecular with one NPTN molecular
| Entire | Name: complex of one PMCA1 molecular with one NPTN molecular |
|---|---|
| Components |
|
-Supramolecule #1: complex of one PMCA1 molecular with one NPTN molecular
| Supramolecule | Name: complex of one PMCA1 molecular with one NPTN molecular type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Plasma membrane calcium-transporting ATPase 1
| Macromolecule | Name: Plasma membrane calcium-transporting ATPase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Ca2+-transporting ATPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 140.987844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDMANNSVA YSGVKNSLKE ANHDGDFGIT LAELRALMEL RSTDALRKIQ ESYGDVYGIC TKLKTSPNEG LSGNPADLER REAVFGKNF IPPKKPKTFL QLVWEALQDV TLIILEIAAI VSLGLSFYQP PEGDNALCGE VSVGEEEGEG ETGWIEGAAI L LSVVCVVL ...String: MGDMANNSVA YSGVKNSLKE ANHDGDFGIT LAELRALMEL RSTDALRKIQ ESYGDVYGIC TKLKTSPNEG LSGNPADLER REAVFGKNF IPPKKPKTFL QLVWEALQDV TLIILEIAAI VSLGLSFYQP PEGDNALCGE VSVGEEEGEG ETGWIEGAAI L LSVVCVVL VTAFNDWSKE KQFRGLQSRI EQEQKFTVIR GGQVIQIPVA DITVGDIAQV KYGDLLPADG ILIQGNDLKI DE SSLTGES DHVKKSLDKD PLLLSGTHVM EGSGRMVVTA VGVNSQTGII FTLLGAGGEE EEKKDEKKKE KKNKKQDGAI ENR NKAKAQ DGAAMEMQPL KSEEGGDGDE KDKKKANLPK KEKSVLQGKL TKLAVQIGKA GLLMSAITVI ILVLYFVIDT FWVQ KRPWL AECTPIYIQY FVKFFIIGVT VLVVAVPEGL PLAVTISLAY SVKKMMKDNN LVRHLDACET MGNATAICSD KTGTL TMNR MTVVQAYINE KHYKKVPEPE AIPPNILSYL VTGISVNCAY TSKILPPEKE GGLPRHVGNK TECALLGLLL DLKRDY QDV RNEIPEEALY KVYTFNSVRK SMSTVLKNSD GSYRIFSKGA SEIILKKCFK ILSANGEAKV FRPRDRDDIV KTVIEPM AS EGLRTICLAF RDFPAGEPEP EWDNENDIVT GLTCIAVVGI EDPVRPEVPD AIKKCQRAGI TVRMVTGDNI NTARAIAT K CGILHPGEDF LCLEGKDFNR RIRNEKGEIE QERIDKIWPK LRVLARSSPT DKHTLVKGII DSTVSDQRQV VAVTGDGTN DGPALKKADV GFAMGIAGTD VAKEASDIIL TDDNFTSIVK AVMWGRNVYD SISKFLQFQL TVNVVAVIVA FTGACITQDS PLKAVQMLW VNLIMDTLAS LALATEPPTE SLLLRKPYGR NKPLISRTMM KNILGHAFYQ LVVVFTLLFA GEKFFDIDSG R NAPLHAPP SEHYTIVFNT FVLMQLFNEI NARKIHGERN VFEGIFNNAI FCTIVLGTFV VQIIIVQFGG KPFSCSELSI EQ WLWSIFL GMGTLLWGQL ISTIPTSRLK FLKEAGHGTQ KEEIPEEELA EDVEEIDHAE RELRRGQILW FRGLNRIQTQ MDV VNAFQS GSSIQGALRR QPSIASQHHD VTNISTPTHI RVVNAFRSSL YEGLEKPESR SSIHNFMTHP EFRIEDSEPH IPLI DDTDA EDDAPTKRNS SPPPSPNKNN NAVDSGIHLT IEMNKSATSS SPGSPLHSLE TSLHHHHHHL EDYKDDDDK UniProtKB: Plasma membrane calcium-transporting ATPase 1 |
-Macromolecule #2: Neuroplastin
| Macromolecule | Name: Neuroplastin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.3284 KDa |
| Sequence | String: MSGSSLPSAL ALSLLLVSGS LLPGPGAAQN EPRIVTSEEV IIRDSPVLPV TLQCNLTSSS HTLTYSYWTK NGVELSATRK NASNMEYRI NKPRAEDSGE YHCVYHFVSA PKANATIEVK AAPDITGHKR SENKNEGQDA TMYCKSVGYP HPDWIWRKKE N GMPMDIVN ...String: MSGSSLPSAL ALSLLLVSGS LLPGPGAAQN EPRIVTSEEV IIRDSPVLPV TLQCNLTSSS HTLTYSYWTK NGVELSATRK NASNMEYRI NKPRAEDSGE YHCVYHFVSA PKANATIEVK AAPDITGHKR SENKNEGQDA TMYCKSVGYP HPDWIWRKKE N GMPMDIVN TSGRFFIINK ENYTELNIVN LQITEDPGEY ECNATNAIGS ASVVTVLRVR SHLAPLWPFL GILAEIIILV VI IVVYEKR KRPDEVPDDD EPAGPMKTNS TNNHKDKNLR QRNTN UniProtKB: Neuroplastin |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)