[English] 日本語
 Yorodumi
Yorodumi- EMDB-6952: Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6952 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationpre-B cell differentiation / Miscellaneous transport and binding events / aspartate transmembrane transport / volume-sensitive anion channel activity / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / monoatomic anion transport ...pre-B cell differentiation / Miscellaneous transport and binding events / aspartate transmembrane transport / volume-sensitive anion channel activity / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / monoatomic anion transport / cell volume homeostasis / response to osmotic stress / intracellular glucose homeostasis / monoatomic ion channel complex / positive regulation of myoblast differentiation / chloride transmembrane transport / positive regulation of insulin secretion / spermatogenesis / intracellular signal transduction / lysosomal membrane / cell surface / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
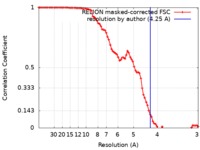
| Method | single particle reconstruction / cryo EM / Resolution: 4.25 Å | |||||||||
 Authors Authors | Kasuya G / Nakane T | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Cryo-EM structures of the human volume-regulated anion channel LRRC8. Authors: Go Kasuya / Takanori Nakane / Takeshi Yokoyama / Yanyan Jia / Masato Inoue / Kengo Watanabe / Ryoki Nakamura / Tomohiro Nishizawa / Tsukasa Kusakizako / Akihisa Tsutsumi / Haruaki Yanagisawa ...Authors: Go Kasuya / Takanori Nakane / Takeshi Yokoyama / Yanyan Jia / Masato Inoue / Kengo Watanabe / Ryoki Nakamura / Tomohiro Nishizawa / Tsukasa Kusakizako / Akihisa Tsutsumi / Haruaki Yanagisawa / Naoshi Dohmae / Motoyuki Hattori / Hidenori Ichijo / Zhiqiang Yan / Masahide Kikkawa / Mikako Shirouzu / Ryuichiro Ishitani / Osamu Nureki /    Abstract: Maintenance of cell volume against osmotic change is crucial for proper cell functions. Leucine-rich repeat-containing 8 proteins are anion-selective channels that extrude anions to decrease the cell ...Maintenance of cell volume against osmotic change is crucial for proper cell functions. Leucine-rich repeat-containing 8 proteins are anion-selective channels that extrude anions to decrease the cell volume on cellular swelling. Here, we present the structure of human leucine-rich repeat-containing 8A, determined by single-particle cryo-electron microscopy. The structure shows a hexameric assembly, and the transmembrane region features a topology similar to gap junction channels. The LRR region, with 15 leucine-rich repeats, forms a long, twisted arc. The channel pore is located along the central axis and constricted on the extracellular side, where highly conserved polar and charged residues at the tip of the extracellular helix contribute to permeability to anions and other osmolytes. Two structural populations were identified, corresponding to compact and relaxed conformations. Comparing the two conformations suggests that the LRR region is flexible and mobile, with rigid-body motions, which might be implicated in structural transitions on pore opening. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6952.map.gz emd_6952.map.gz | 48.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6952-v30.xml emd-6952-v30.xml emd-6952.xml emd-6952.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6952_fsc.xml emd_6952_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_6952.png emd_6952.png | 169.7 KB | ||
| Masks |  emd_6952_msk_1.map emd_6952_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-6952.cif.gz emd-6952.cif.gz | 6.5 KB | ||
| Others |  emd_6952_half_map_1.map.gz emd_6952_half_map_1.map.gz emd_6952_half_map_2.map.gz emd_6952_half_map_2.map.gz | 49.4 MB 49.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6952 http://ftp.pdbj.org/pub/emdb/structures/EMD-6952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6952 | HTTPS FTP |
-Related structure data
| Related structure data |  5zsuMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10196 (Title: CryoEM structure of human LRRC8A / Data size: 2.9 TB EMPIAR-10196 (Title: CryoEM structure of human LRRC8A / Data size: 2.9 TBData #1: Unaligned movies of human LRRC8A [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6952.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6952.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.49 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
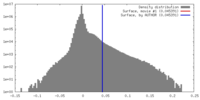
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_6952_msk_1.map emd_6952_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_6952_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_6952_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric channel of LRC8A_HUMAN
| Entire | Name: Hexameric channel of LRC8A_HUMAN |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric channel of LRC8A_HUMAN
| Supramolecule | Name: Hexameric channel of LRC8A_HUMAN / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Volume-regulated anion channel subunit LRRC8A
| Macromolecule | Name: Volume-regulated anion channel subunit LRRC8A / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 95.271516 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIPVTELRYF ADTQPAYRIL KPWWDVFTDY ISIVMLMIAV FGGTLQVTQD KMICLPCKWV TKDSCNDSFR GWAAPGPEPT YPNSTILPT PDTGPTGIKY DLDRHQYNYV DAVCYENRLH WFAKYFPYLV LLHTLIFLAC SNFWFKFPRT SSKLEHFVSI L LKCFDSPW ...String: MIPVTELRYF ADTQPAYRIL KPWWDVFTDY ISIVMLMIAV FGGTLQVTQD KMICLPCKWV TKDSCNDSFR GWAAPGPEPT YPNSTILPT PDTGPTGIKY DLDRHQYNYV DAVCYENRLH WFAKYFPYLV LLHTLIFLAC SNFWFKFPRT SSKLEHFVSI L LKCFDSPW TTRALSETVV EESDPKPAFS KMNGSMDKKS STVSEDVEAT VPMLQRTKSR IEQGIVDRSE TGVLDKKEGE QA KALFEKV KKFRTHVEEG DIVYRLYMRQ TIIKVIKFIL IICYTVYYVH NIKFDVDCTV DIESLTGYRT YRCAHPLATL FKI LASFYI SLVIFYGLIC MYTLWWMLRR SLKKYSFESI REESSYSDIP DVKNDFAFML HLIDQYDPLY SKRFAVFLSE VSEN KLRQL NLNNEWTLDK LRQRLTKNAQ DKLELHLFML SGIPDTVFDL VELEVLKLEL IPDVTIPPSI AQLTGLKELW LYHTA AKIE APALAFLREN LRALHIKFTD IKEIPLWIYS LKTLEELHLT GNLSAENNRY IVIDGLRELK RLKVLRLKSN LSKLPQ VVT DVGVHLQKLS INNEGTKLIV LNSLKKMANL TELELIRCDL ERIPHSIFSL HNLQEIDLKD NNLKTIEEII SFQHLHR LT CLKLWYNHIA YIPIQIGNLT NLERLYLNRN KIEKIPTQLF YCRKLRYLDL SHNNLTFLPA DIGLLQNLQN LAITANRI E TLPPELFQCR KLRALHLGNN VLQSLPSRVG ELTNLTQIEL RGNRLECLPV ELGECPLLKR SGLVVEEDLF NTLPPEVKE RLWRADKEQA GTENLYFQ UniProtKB: Volume-regulated anion channel subunit LRRC8A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Solution was freshly prepared to avoid digitonin precipitation. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER/RHODIUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 53.329 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blotted for 12 seconds before plunging.. | |||||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 79.55 K / Max: 79.55 K |
| Details | Specimen holder is FEI Talos Arctica autogrid holder. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 3 / Number real images: 5305 / Average exposure time: 15.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 23500 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-5zsu: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)