Entry Database : EMDB / ID : EMD-6864Title The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution Complex : Human Catalytic Step I SpliceosomeProtein or peptide : x 41 typesRNA : x 4 typesLigand : x 6 types / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / Resolution : 4.1 Å Zhan X / Yan C Funding support Organization Grant number Country Ministry of Science and Technology (China) 2014ZX09507003006 National Natural Science Foundation of China 31130002 National Natural Science Foundation of China 31321062
Journal : Science / Year : 2018Title : Structure of a human catalytic step I spliceosome.Authors : Xiechao Zhan / Chuangye Yan / Xiaofeng Zhang / Jianlin Lei / Yigong Shi / Abstract : Splicing by the spliceosome involves branching and exon ligation. The branching reaction leads to the formation of the catalytic step I spliceosome (C complex). Here we report the cryo-electron ... Splicing by the spliceosome involves branching and exon ligation. The branching reaction leads to the formation of the catalytic step I spliceosome (C complex). Here we report the cryo-electron microscopy structure of the human C complex at an average resolution of 4.1 angstroms. Compared with the C complex, the human complex contains 11 additional proteins. The step I splicing factors CCDC49 and CCDC94 (Cwc25 and Yju2 in , respectively) closely interact with the DEAH-family adenosine triphosphatase/helicase Prp16 and bridge the gap between Prp16 and the active-site RNA elements. These features, together with structural comparison of the human C and C* complexes, provide mechanistic insights into ribonucleoprotein remodeling and allow the proposition of a working mechanism for the C-to-C* transition. History Deposition Dec 14, 2017 - Header (metadata) release Aug 8, 2018 - Map release Aug 8, 2018 - Update Oct 23, 2024 - Current status Oct 23, 2024 Processing site : PDBj / Status : Released
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Map data
Map data Sample
Sample Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors China, 3 items
China, 3 items  Citation
Citation Journal: Science / Year: 2018
Journal: Science / Year: 2018
 Structure visualization
Structure visualization Movie viewer
Movie viewer SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links emd_6864.map.gz
emd_6864.map.gz EMDB map data format
EMDB map data format emd-6864-v30.xml
emd-6864-v30.xml emd-6864.xml
emd-6864.xml EMDB header
EMDB header emd_6864.png
emd_6864.png emd-6864.cif.gz
emd-6864.cif.gz emd_6864_additional.map.gz
emd_6864_additional.map.gz emd_6864_half_map_1.map.gz
emd_6864_half_map_1.map.gz emd_6864_half_map_2.map.gz
emd_6864_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-6864
http://ftp.pdbj.org/pub/emdb/structures/EMD-6864 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6864
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6864
 Links
Links EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource Map
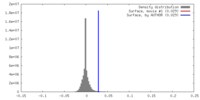
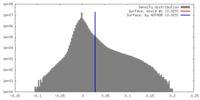
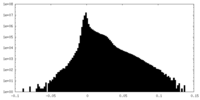
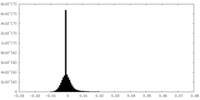
Map Download / File: emd_6864.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Download / File: emd_6864.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Sample components
Sample components Processing
Processing Sample preparation
Sample preparation Electron microscopy
Electron microscopy FIELD EMISSION GUN
FIELD EMISSION GUN
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)