+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6835 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mononucleosome from HP1alpha-dinucleosome-H3K9me3 | ||||||||||||
 Map data Map data | mononucleosome reconstructed from_HP1a-dinucleosome-H3K9me3 data | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
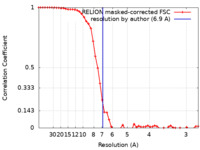
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | ||||||||||||
 Authors Authors | Machida S / Takizawa Y / Ishimaru M / Sekine S / Sugita Y / Nakayama J / Wolf M / Kurumizaka H | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2018 Journal: Mol Cell / Year: 2018Title: Structural Basis of Heterochromatin Formation by Human HP1. Authors: Shinichi Machida / Yoshimasa Takizawa / Masakazu Ishimaru / Yukihiko Sugita / Satoshi Sekine / Jun-Ichi Nakayama / Matthias Wolf / Hitoshi Kurumizaka /  Abstract: Heterochromatin plays important roles in transcriptional silencing and genome maintenance by the formation of condensed chromatin structures, which determine the epigenetic status of eukaryotic cells. ...Heterochromatin plays important roles in transcriptional silencing and genome maintenance by the formation of condensed chromatin structures, which determine the epigenetic status of eukaryotic cells. The trimethylation of histone H3 lysine 9 (H3K9me3), a target of heterochromatin protein 1 (HP1), is a hallmark of heterochromatin formation. However, the mechanism by which HP1 folds chromatin-containing H3K9me3 into a higher-order structure has not been elucidated. Here we report the three-dimensional structure of the H3K9me3-containing dinucleosomes complexed with human HP1α, HP1β, and HP1γ, determined by cryogenic electron microscopy with a Volta phase plate. In the structures, two H3K9me3 nucleosomes are bridged by a symmetric HP1 dimer. Surprisingly, the linker DNA between the nucleosomes does not directly interact with HP1, thus allowing nucleosome remodeling by the ATP-utilizing chromatin assembly and remodeling factor (ACF). The structure depicts the fundamental architecture of heterochromatin. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6835.map.gz emd_6835.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6835-v30.xml emd-6835-v30.xml emd-6835.xml emd-6835.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6835_fsc.xml emd_6835_fsc.xml | 5.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_6835.png emd_6835.png | 57.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6835 http://ftp.pdbj.org/pub/emdb/structures/EMD-6835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6835 | HTTPS FTP |
-Validation report
| Summary document |  emd_6835_validation.pdf.gz emd_6835_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6835_full_validation.pdf.gz emd_6835_full_validation.pdf.gz | 78 KB | Display | |
| Data in XML |  emd_6835_validation.xml.gz emd_6835_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6835 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6835 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6835 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6835 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6835.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6835.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mononucleosome reconstructed from_HP1a-dinucleosome-H3K9me3 data | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mononucleosome from HP1alpha-dinucleosome-H3K9me3
| Entire | Name: Mononucleosome from HP1alpha-dinucleosome-H3K9me3 |
|---|---|
| Components |
|
-Supramolecule #1: Mononucleosome from HP1alpha-dinucleosome-H3K9me3
| Supramolecule | Name: Mononucleosome from HP1alpha-dinucleosome-H3K9me3 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Recombinant plasmid: pH2A, pH2B, pH3.2K9C/C110A, pH4, pHP1a-pre |
| Molecular weight | Theoretical: 500 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | .8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 100.0 nm / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 9.33 kPa / Details: Gatan Solarus | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV / Details: 3 second blot, 2.5uL. | |||||||||
| Details | sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 100.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.1 mrad |
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: -10 eV / Energy filter - Upper energy threshold: 10 eV |
| Details | nanoprobe, parallel beam illumination |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-40 / Number grids imaged: 2 / Number real images: 2329 / Average exposure time: 5.0 sec. / Average electron dose: 38.0 e/Å2 Details: Images were collected with a volta phase plate (VPP) in-focus |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 35971 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.62 mm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)