+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6657 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a eukaryotic cyclic nucleotide-gated channel | |||||||||
 Map data Map data | TAX-4 density map without symmetry imposed after post-processing in RELION1.4. The reported resolution is 4.5A. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Li X | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structure of a eukaryotic cyclic-nucleotide-gated channel. Authors: Minghui Li / Xiaoyuan Zhou / Shu Wang / Ioannis Michailidis / Ye Gong / Deyuan Su / Huan Li / Xueming Li / Jian Yang /   Abstract: Cyclic-nucleotide-gated channels are essential for vision and olfaction. They belong to the voltage-gated ion channel superfamily but their activities are controlled by intracellular cyclic ...Cyclic-nucleotide-gated channels are essential for vision and olfaction. They belong to the voltage-gated ion channel superfamily but their activities are controlled by intracellular cyclic nucleotides instead of transmembrane voltage. Here we report a 3.5-Å-resolution single-particle electron cryo-microscopy structure of a cyclic-nucleotide-gated channel from Caenorhabditis elegans in the cyclic guanosine monophosphate (cGMP)-bound open state. The channel has an unusual voltage-sensor-like domain, accounting for its deficient voltage dependence. A carboxy-terminal linker connecting S6 and the cyclic-nucleotide-binding domain interacts directly with both the voltage-sensor-like domain and the pore domain, forming a gating ring that couples conformational changes triggered by cyclic nucleotide binding to the gate. The selectivity filter is lined by the carboxylate side chains of a functionally important glutamate and three rings of backbone carbonyls. This structure provides a new framework for understanding mechanisms of ion permeation, gating and channelopathy of cyclic-nucleotide-gated channels and cyclic nucleotide modulation of related channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6657.map.gz emd_6657.map.gz | 2.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6657-v30.xml emd-6657-v30.xml emd-6657.xml emd-6657.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6657.png emd_6657.png | 139.5 KB | ||
| Others |  emd_6657_additional.map.gz emd_6657_additional.map.gz | 11.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6657 http://ftp.pdbj.org/pub/emdb/structures/EMD-6657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6657 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6657.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6657.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TAX-4 density map without symmetry imposed after post-processing in RELION1.4. The reported resolution is 4.5A. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
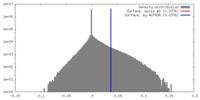
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: TAX-4 density map without symmetry imposed before post-processing...
| File | emd_6657_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TAX-4 density map without symmetry imposed before post-processing in RELION1.4. The reported resolution is 5.1A. | ||||||||||||
| Projections & Slices |
| ||||||||||||
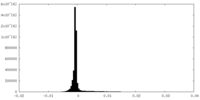
| Density Histograms |
- Sample components
Sample components
-Entire : Cyclic nucleotide-gated (CNG) channels
| Entire | Name: Cyclic nucleotide-gated (CNG) channels |
|---|---|
| Components |
|
-Supramolecule #1: Cyclic nucleotide-gated (CNG) channels
| Supramolecule | Name: Cyclic nucleotide-gated (CNG) channels / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) / Recombinant plasmid: pFastBac1 Trichoplusia ni (cabbage looper) / Recombinant plasmid: pFastBac1 |
-Macromolecule #1: Cyclic nucleotide-gated (CNG) channels
| Macromolecule | Name: Cyclic nucleotide-gated (CNG) channels / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GGGGSMSTAE PAPDPTNPST SGLAPTTNGI GSPPPTASAA TKFSILTKFL RRKNQVHTTT AQQNEFMQKY MPNGNSNAVQ PAATGGQPAS SDGGSAIEVP PPKESYAVRI RKYLANYTQD PSTDNFYYWT CVVTVAYIYN LLFVIARQVF NDLIGPSSQS LCRFYNGTLN ...String: GGGGSMSTAE PAPDPTNPST SGLAPTTNGI GSPPPTASAA TKFSILTKFL RRKNQVHTTT AQQNEFMQKY MPNGNSNAVQ PAATGGQPAS SDGGSAIEVP PPKESYAVRI RKYLANYTQD PSTDNFYYWT CVVTVAYIYN LLFVIARQVF NDLIGPSSQS LCRFYNGTLN STTQVECTYN MLTNMKEMPT YSQYPDLGWS KYWHFRMLWV FFDLLMDCVY LIDTFLNYRM GYMDQGLVVR EAEKVTKAYW QSKQYRIDGI SLIPLDYILG WPIPYINWRG LPILRLNRLI RYKRVRNCLE RTETRSSMPN AFRVVVVVWY IVIIIHWNAC LYFWISEWIG LGTDAWVYGH LNKQSLPDDI TDTLLRRYVY SFYWSTLILT TIGEVPSPVR NIEYAFVTLD LMCGVLIFAT IVGNVGSMIS NMSAARTEFQ NKMDGIKQYM ELRKVSKQLE IRVIKWFDYL WTNKQSLSDQ QVLKVLPDKL QAEIAMQVHF ETLRKVRIFQ DCEAGLLAEL VLKLQLQVFS PGDFICKKGD IGREMYIVKR GRLQVVDDDG KKVFVTLQEG SVFGELSILN IAGSKNGNRR TANVRSVGYT DLFVLSKTDL WNALREYPDA RKLLLAKGRE ILKKDNLLDE NAPEEQKTVE EIAEHLNNAV KVLQTRMARL IVEHSSTEGK LMKRIEMLEK HLSRYKALAR RQKTMHGVSI DGGDISTDGV DERVRPPRLR QTKTIDLPTG TESESLLK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV Details: Blotted for 4.0 seconds(double-sided, blot force 1),after waiting for 3 seconds the grid was immediately plunged into liquid ethane cooled by liquid-nitrogen.. | |||||||||||||||
| Details | this sample was homogeneous |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-32 / Average exposure time: 8.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)