+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6465 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Architecture of a Eukaryotic Replisome | |||||||||
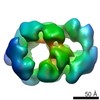
 Map data Map data | Yeast CMGE complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Eukaryotic replisome / CMGE / Electron microscopy | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 16.0 Å | |||||||||
 Authors Authors | Sun J / Shi Y / Georgescu RE / Yuan Z / Chait B / Li H / O'Donnell ME | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2015 Journal: Nat Struct Mol Biol / Year: 2015Title: The architecture of a eukaryotic replisome. Authors: Jingchuan Sun / Yi Shi / Roxana E Georgescu / Zuanning Yuan / Brian T Chait / Huilin Li / Michael E O'Donnell /  Abstract: At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase is positioned in front to unwind DNA and that DNA polymerases trail behind the helicase. Here ...At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase is positioned in front to unwind DNA and that DNA polymerases trail behind the helicase. Here we used single-particle EM to directly image a Saccharomyces cerevisiae replisome. Contrary to expectations, the leading strand Pol ɛ is positioned ahead of CMG helicase, whereas Ctf4 and the lagging-strand polymerase (Pol) α-primase are behind the helicase. This unexpected architecture indicates that the leading-strand DNA travels a long distance before reaching Pol ɛ, first threading through the Mcm2-7 ring and then making a U-turn at the bottom and reaching Pol ɛ at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture and provides a basis for further structural and biochemical replisome studies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6465.map.gz emd_6465.map.gz | 58.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6465-v30.xml emd-6465-v30.xml emd-6465.xml emd-6465.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6465.jpg emd_6465.jpg | 21.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6465 http://ftp.pdbj.org/pub/emdb/structures/EMD-6465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6465 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6465.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6465.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast CMGE complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae CMGE complex
| Entire | Name: Saccharomyces cerevisiae CMGE complex |
|---|---|
| Components |
|
-Supramolecule #1000: Saccharomyces cerevisiae CMGE complex
| Supramolecule | Name: Saccharomyces cerevisiae CMGE complex / type: sample / ID: 1000 / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 1.2 MDa |
-Macromolecule #1: CMG complex
| Macromolecule | Name: CMG complex / type: protein_or_peptide / ID: 1 / Name.synonym: CMG / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 700 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: Polymerase Epsilon
| Macromolecule | Name: Polymerase Epsilon / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 500 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | Details: 20 mM Tris-acetate, 40 mM potassium glutamate, 2 mM DTT, 0.1 mM EDTA |
| Staining | Type: NEGATIVE Details: Grids were stained with 1% w/v uranyl acetate for 1 minute. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Date | Sep 10, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 320 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | Particles were selected in EMAN 2.1 using SWAM. |
|---|---|
| CTF correction | Details: CTFFIND3 |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: OTHER / Software - Name: Relion_1.3 / Number images used: 18721 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















