[English] 日本語
 Yorodumi
Yorodumi- EMDB-6891: Cryo-EM structure of a human activated spliceosome (early Bact) a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6891 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | ||||||||||||
 Map data Map data | Cryo-EM structure of the human activated spliceosome (early Bact) at 4.8 angstrom | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | spliceosome / cryo-EM structure / activated spliceosome / early Bact complex / pre-mRNA splicing / SPLICING | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationRES complex / negative regulation of chemokine-mediated signaling pathway / snoRNA splicing / U11/U12 snRNP / regulation of retinoic acid receptor signaling pathway / post-mRNA release spliceosomal complex / U2 snRNP binding / U7 snRNA binding / histone pre-mRNA DCP binding / U7 snRNP ...RES complex / negative regulation of chemokine-mediated signaling pathway / snoRNA splicing / U11/U12 snRNP / regulation of retinoic acid receptor signaling pathway / post-mRNA release spliceosomal complex / U2 snRNP binding / U7 snRNA binding / histone pre-mRNA DCP binding / U7 snRNP / cis assembly of pre-catalytic spliceosome / histone pre-mRNA 3'end processing complex / regulation of vitamin D receptor signaling pathway / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / spliceosome conformational change to release U4 (or U4atac) and U1 (or U11) / B-WICH complex / miRNA processing / nuclear retinoic acid receptor binding / protein methylation / U12-type spliceosomal complex / 7-methylguanosine cap hypermethylation / U1 snRNP binding / U2-type catalytic step 1 spliceosome / RNA splicing, via transesterification reactions / methylosome / pICln-Sm protein complex / positive regulation of mRNA splicing, via spliceosome / regulation of mRNA splicing, via spliceosome / snRNP binding / sno(s)RNA-containing ribonucleoprotein complex / blastocyst formation / small nuclear ribonucleoprotein complex / splicing factor binding / Notch binding / SMN-Sm protein complex / spliceosomal tri-snRNP complex / host-mediated activation of viral transcription / mRNA cis splicing, via spliceosome / U2-type precatalytic spliceosome / P granule / positive regulation of vitamin D receptor signaling pathway / commitment complex / telomerase holoenzyme complex / nuclear vitamin D receptor binding / U2-type prespliceosome assembly / U2-type spliceosomal complex / Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells / telomerase RNA binding / RUNX3 regulates NOTCH signaling / U2-type catalytic step 2 spliceosome / NOTCH4 Intracellular Domain Regulates Transcription / SAGA complex / U2 snRNP / U1 snRNP / RNA Polymerase II Transcription Termination / U4 snRNP / NOTCH3 Intracellular Domain Regulates Transcription / positive regulation of neurogenesis / U2-type prespliceosome / positive regulation of transcription by RNA polymerase III / K63-linked polyubiquitin modification-dependent protein binding / nuclear androgen receptor binding / precatalytic spliceosome / WD40-repeat domain binding / Notch-HLH transcription pathway / Formation of paraxial mesoderm / pattern recognition receptor activity / SMAD binding / positive regulation of transforming growth factor beta receptor signaling pathway / regulation of RNA splicing / spliceosomal complex assembly / mRNA 3'-splice site recognition / positive regulation of transcription by RNA polymerase I / mRNA Splicing - Minor Pathway / spliceosomal tri-snRNP complex assembly / Prp19 complex / U5 snRNA binding / U5 snRNP / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / U2 snRNA binding / positive regulation of G1/S transition of mitotic cell cycle / U6 snRNA binding / pre-mRNA intronic binding / protein localization to nucleus / spliceosomal snRNP assembly / U1 snRNA binding / RNA processing / Cajal body / regulation of DNA repair / transcription regulator inhibitor activity / retinoic acid receptor signaling pathway / U4/U6 x U5 tri-snRNP complex / cellular response to retinoic acid / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / antiviral innate immune response / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / RNA splicing / negative regulation of canonical NF-kappaB signal transduction Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  unidentified adenovirus unidentified adenovirus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | ||||||||||||
 Authors Authors | Zhang X / Yan C | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2018 Journal: Cell Res / Year: 2018Title: Structure of the human activated spliceosome in three conformational states. Authors: Xiaofeng Zhang / Chuangye Yan / Xiechao Zhan / Lijia Li / Jianlin Lei / Yigong Shi /  Abstract: During each cycle of pre-mRNA splicing, the pre-catalytic spliceosome (B complex) is converted into the activated spliceosome (B complex), which has a well-formed active site but cannot proceed to ...During each cycle of pre-mRNA splicing, the pre-catalytic spliceosome (B complex) is converted into the activated spliceosome (B complex), which has a well-formed active site but cannot proceed to the branching reaction. Here, we present the cryo-EM structure of the human B complex in three distinct conformational states. The EM map allows atomic modeling of nearly all protein components of the U2 small nuclear ribonucleoprotein (snRNP), including three of the SF3a complex and seven of the SF3b complex. The structure of the human B complex contains 52 proteins, U2, U5, and U6 small nuclear RNA (snRNA), and a pre-mRNA. Three distinct conformations have been captured, representing the early, mature, and late states of the human B complex. These complexes differ in the orientation of the Switch loop of Prp8, the splicing factors RNF113A and NY-CO-10, and most components of the NineTeen complex (NTC) and the NTC-related complex. Analysis of these three complexes and comparison with the B and C complexes reveal an ordered flux of components in the B-to-B and the B-to-B transitions, which ultimately prime the active site for the branching reaction. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6891.map.gz emd_6891.map.gz | 226.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6891-v30.xml emd-6891-v30.xml emd-6891.xml emd-6891.xml | 64.9 KB 64.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6891.png emd_6891.png | 163.3 KB | ||
| Filedesc metadata |  emd-6891.cif.gz emd-6891.cif.gz | 20.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6891 http://ftp.pdbj.org/pub/emdb/structures/EMD-6891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6891 | HTTPS FTP |
-Related structure data
| Related structure data |  5z58MC  6889C  6890C  5z56C  5z57C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6891.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6891.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the human activated spliceosome (early Bact) at 4.8 angstrom | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.338 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
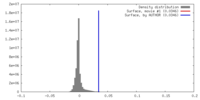
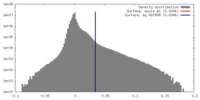
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : early Bact spliceosome
+Supramolecule #1: early Bact spliceosome
+Macromolecule #1: Pre-mRNA-processing-splicing factor 8
+Macromolecule #3: 116 kDa U5 small nuclear ribonucleoprotein component
+Macromolecule #4: U5 small nuclear ribonucleoprotein 200 kDa helicase
+Macromolecule #5: U5 small nuclear ribonucleoprotein 40 kDa protein
+Macromolecule #6: Small nuclear ribonucleoprotein Sm D3
+Macromolecule #7: Small nuclear ribonucleoprotein-associated proteins B and B'
+Macromolecule #8: Small nuclear ribonucleoprotein Sm D1
+Macromolecule #9: Small nuclear ribonucleoprotein Sm D2
+Macromolecule #10: Small nuclear ribonucleoprotein F
+Macromolecule #11: Small nuclear ribonucleoprotein E
+Macromolecule #12: Small nuclear ribonucleoprotein G
+Macromolecule #16: U2 small nuclear ribonucleoprotein A'
+Macromolecule #17: U2 small nuclear ribonucleoprotein B''
+Macromolecule #18: Splicing factor 3A subunit 3
+Macromolecule #19: Splicing factor 3A subunit 1
+Macromolecule #20: Splicing factor 3A subunit 2
+Macromolecule #21: Splicing factor 3B subunit 1
+Macromolecule #22: Splicing factor 3B subunit 2
+Macromolecule #23: Splicing factor 3B subunit 3
+Macromolecule #24: Splicing factor 3B subunit 4
+Macromolecule #25: Splicing factor 3B subunit 6
+Macromolecule #26: PHD finger-like domain-containing protein 5A
+Macromolecule #27: Splicing factor 3B subunit 5
+Macromolecule #28: Crooked neck-like protein 1
+Macromolecule #29: Cell division cycle 5-like protein
+Macromolecule #30: RING finger protein 113A
+Macromolecule #31: Spliceosome-associated protein CWC15 homolog
+Macromolecule #32: Skip
+Macromolecule #33: Pleiotropic regulator 1
+Macromolecule #34: Pre-mRNA-splicing factor CWC22 homolog
+Macromolecule #35: Smad nuclear-interacting protein 1
+Macromolecule #36: RNA-binding motif protein, X-linked 2
+Macromolecule #37: BUD13 homolog
+Macromolecule #38: Peptidyl-prolyl cis-trans isomerase CWC27 homolog
+Macromolecule #39: Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16
+Macromolecule #2: U5 snRNA
+Macromolecule #13: U6 snRNA
+Macromolecule #14: pre-mRNA
+Macromolecule #15: U2 snRNA
+Macromolecule #40: INOSITOL HEXAKISPHOSPHATE
+Macromolecule #41: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #42: MAGNESIUM ION
+Macromolecule #43: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 / Details: 20 mM HEPES-KOH, pH 7.9, 150 mM NaCl, 1.5 mM MgCl2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)