[English] 日本語
 Yorodumi
Yorodumi- EMDB-6366: The cryoEM map of EV71 mature viron in complex with the Fab fragm... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6366 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | |||||||||
 Map data Map data | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Enterovirus 71(EV71) / virus-antibody complex / bivalent binding / high resolution cryo-EM | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / virion component / ribonucleoside triphosphate phosphatase activity / nucleoside-triphosphate phosphatase ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / virion component / ribonucleoside triphosphate phosphatase activity / nucleoside-triphosphate phosphatase / host cell / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / symbiont entry into host cell / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human enterovirus 71 Human enterovirus 71 | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 4.8 Å | |||||||||
 Authors Authors | Fan C / Ye XH / Ku ZQ / Zuo T / Kong LL / Zhang C / Shi JP / Liu QW / Chen T / Zhang YY ...Fan C / Ye XH / Ku ZQ / Zuo T / Kong LL / Zhang C / Shi JP / Liu QW / Chen T / Zhang YY / Jiang W / Zhang LQ / Huang Z / Cong Y | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2016 Journal: PLoS Pathog / Year: 2016Title: Structural Basis for Recognition of Human Enterovirus 71 by a Bivalent Broadly Neutralizing Monoclonal Antibody. Authors: Xiaohua Ye / Chen Fan / Zhiqiang Ku / Teng Zuo / Liangliang Kong / Chao Zhang / Jinping Shi / Qingwei Liu / Tan Chen / Yingyi Zhang / Wen Jiang / Linqi Zhang / Zhong Huang / Yao Cong /   Abstract: Enterovirus 71 (EV71) is the main pathogen responsible for hand, foot and mouth disease with severe neurological complications and even death in young children. We have recently identified a highly ...Enterovirus 71 (EV71) is the main pathogen responsible for hand, foot and mouth disease with severe neurological complications and even death in young children. We have recently identified a highly potent anti-EV71 neutralizing monoclonal antibody, termed D5. Here we investigated the structural basis for recognition of EV71 by the antibody D5. Four three-dimensional structures of EV71 particles in complex with IgG or Fab of D5 were reconstructed by cryo-electron microscopy (cryo-EM) single particle analysis all at subnanometer resolutions. The most critical EV71 mature virion-Fab structure was resolved to a resolution of 4.8 Å, which is rare in cryo-EM studies of virus-antibody complex so far. The structures reveal a bivalent binding pattern of D5 antibody across the icosahedral 2-fold axis on mature virion, suggesting that D5 binding may rigidify virions to prevent their conformational changes required for subsequent RNA release. Moreover, we also identified that the complementary determining region 3 (CDR3) of D5 heavy chain directly interacts with the extremely conserved VP1 GH-loop of EV71, which was validated by biochemical and virological assays. We further showed that D5 is indeed able to neutralize a variety of EV71 genotypes and strains. Moreover, D5 could potently confer protection in a mouse model of EV71 infection. Since the conserved VP1 GH-loop is involved in EV71 binding with its uncoating receptor, the scavenger receptor class B, member 2 (SCARB2), the broadly neutralizing ability of D5 might attribute to its inhibition of EV71 from binding SCARB2. Altogether, our results elucidate the structural basis for the binding and neutralization of EV71 by the broadly neutralizing antibody D5, thereby enhancing our understanding of antibody-based protection against EV71 infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6366.map.gz emd_6366.map.gz | 113.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6366-v30.xml emd-6366-v30.xml emd-6366.xml emd-6366.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6366.jpg emd_6366.jpg | 149.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6366 http://ftp.pdbj.org/pub/emdb/structures/EMD-6366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6366 | HTTPS FTP |
-Related structure data
| Related structure data |  3jauMC  6365C  6383C  6384C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6366.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6366.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.79 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
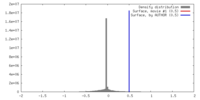
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EV71 mature viron in complex with the Fab fragment of antibody D5
| Entire | Name: EV71 mature viron in complex with the Fab fragment of antibody D5 |
|---|---|
| Components |
|
-Supramolecule #1000: EV71 mature viron in complex with the Fab fragment of antibody D5
| Supramolecule | Name: EV71 mature viron in complex with the Fab fragment of antibody D5 type: sample / ID: 1000 Oligomeric state: One fab fragment of antibody D5 bind to one protomer of EV71 Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 8 MDa |
-Supramolecule #1: Human enterovirus 71
| Supramolecule | Name: Human enterovirus 71 / type: virus / ID: 1 Details: EV71 mature viron in complex with the Fab fragment of antibody D5 NCBI-ID: 39054 / Sci species name: Human enterovirus 71 / Sci species strain: G082 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Molecular weight | Theoretical: 5 MDa |
| Virus shell | Shell ID: 1 / Diameter: 300 Å / T number (triangulation number): 3 |
-Macromolecule #1: antibody D5
| Macromolecule | Name: antibody D5 / type: protein_or_peptide / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 0.15 M phosphate buffered saline(PBS) buffer |
| Staining | Type: NEGATIVE Details: Grids were plunge-frozen into liquid ethane using a FEI Mark IV virtobot, after 2s blotting to remove extra sample. |
| Grid | Details: 200 mesh R1.2x1.3 Quantifoil Cu grid, glow discharged in air. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK IV / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90 K / Max: 92 K / Average: 91 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 75000 magnification |
| Date | Oct 2, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 320 / Average electron dose: 16 e/Å2 Details: Every image is the average of seven frames recorded by the direct electron detector |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.005 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 37000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were boxed using e2boxer.py. CTF fitting was automatically performed using fitctf2.py in jspr, then visually validated and adjusted using EMAN1.9 ctfit program. The gold standard 3D reconstruction procedure was followed using jspr package, with the datasets split into two halves in the beginning. |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.8 Å / Resolution method: OTHER / Software - Name: jspr, EMAN, EMAN2 / Number images used: 2902 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-3jau: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)