+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10127 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thermally-inactivated murine norovirus (MNV-1) | ||||||||||||
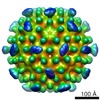
 Map data Map data | EM density map for heat-inactivated (hi)MNV, filtered by local resolution. | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |   Murine norovirus 1 Murine norovirus 1 | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Snowden JS / Hurdiss DL / Adeyemi OO / Ranson NA / Herod MR / Stonehouse NJ | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2020 Journal: PLoS Biol / Year: 2020Title: Dynamics in the murine norovirus capsid revealed by high-resolution cryo-EM. Authors: Joseph S Snowden / Daniel L Hurdiss / Oluwapelumi O Adeyemi / Neil A Ranson / Morgan R Herod / Nicola J Stonehouse /  Abstract: Icosahedral viral capsids must undergo conformational rearrangements to coordinate essential processes during the viral life cycle. Capturing such conformational flexibility has been technically ...Icosahedral viral capsids must undergo conformational rearrangements to coordinate essential processes during the viral life cycle. Capturing such conformational flexibility has been technically challenging yet could be key for developing rational therapeutic agents to combat infections. Noroviruses are nonenveloped, icosahedral viruses of global importance to human health. They are a common cause of acute gastroenteritis, yet no vaccines or specific antiviral agents are available. Here, we use genetics and cryo-electron microscopy (cryo-EM) to study the high-resolution solution structures of murine norovirus as a model for human viruses. By comparing our 3 structures (at 2.9- to 3.1-Å resolution), we show that whilst there is little change to the shell domain of the capsid, the radiating protruding domains are flexible, adopting distinct states both independently and synchronously. In doing so, the capsids sample a range of conformational space, with implications for maintaining virion stability and infectivity. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10127.map.gz emd_10127.map.gz | 184.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10127-v30.xml emd-10127-v30.xml emd-10127.xml emd-10127.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10127_fsc.xml emd_10127_fsc.xml | 15.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_10127.png emd_10127.png | 170.3 KB | ||
| Masks |  emd_10127_msk_1.map emd_10127_msk_1.map | 325 MB |  Mask map Mask map | |
| Others |  emd_10127_additional_1.map.gz emd_10127_additional_1.map.gz emd_10127_additional_2.map.gz emd_10127_additional_2.map.gz emd_10127_half_map_1.map.gz emd_10127_half_map_1.map.gz emd_10127_half_map_2.map.gz emd_10127_half_map_2.map.gz | 255.7 MB 300.4 MB 255.4 MB 255.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10127 http://ftp.pdbj.org/pub/emdb/structures/EMD-10127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10127 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10127.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10127.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM density map for heat-inactivated (hi)MNV, filtered by local resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10127_msk_1.map emd_10127_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened density map from 3D refinement.
| File | emd_10127_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened density map from 3D refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened density map.
| File | emd_10127_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened density map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: First half map from 3D refinement.
| File | emd_10127_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map from 3D refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Second half map from 3D refinement.
| File | emd_10127_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map from 3D refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Murine norovirus 1
| Entire | Name:   Murine norovirus 1 Murine norovirus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Murine norovirus 1
| Supramolecule | Name: Murine norovirus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: RAW264.7 cells / NCBI-ID: 223997 / Sci species name: Murine norovirus 1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / T number (triangulation number): 3 |
-Macromolecule #1: Murine norovirus (MNV-1) VP1
| Macromolecule | Name: Murine norovirus (MNV-1) VP1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Murine norovirus 1 Murine norovirus 1 |
| Sequence | String: MRMSDGAAPK ANGSEASGQD LVPAAVEQAV PIQPVAGAAL AAPAAGQINQ IDPWIFQNFV QCPLGEFSIS PRNTPGEILF DLALGPGLNP YLAHLSAMYT GWVGNMEVQL VLAGNAFTAG KVVVALVPPY FPKGSLTTAQ ITCFPHVMCD VRTLEPIQLP LLDVRRVLWH ...String: MRMSDGAAPK ANGSEASGQD LVPAAVEQAV PIQPVAGAAL AAPAAGQINQ IDPWIFQNFV QCPLGEFSIS PRNTPGEILF DLALGPGLNP YLAHLSAMYT GWVGNMEVQL VLAGNAFTAG KVVVALVPPY FPKGSLTTAQ ITCFPHVMCD VRTLEPIQLP LLDVRRVLWH ATQDQEESMR LVCMLYTPLR TNSPGDESFV VSGRLLSKPA ADFNFVYLTP PIERTIYRMV DLPVIQPRLC THARWPAPVY GLLVDPSLPS NPQWQNGRVH VDGTLLGTTP ISGSWVSCFA AEAAYEFQSG TGEVATFTLI EQDGSAYVPG DRAAPLGYPD FSGQLEIEVQ TETTKTGDKL KVTTFEMILG PTTNADQAPY QGRVFASVTA AASLDLVDGR VRAVPRSIYG FQDTIPEYND GLLVPLAPPI GPFLPGEVLL RFRTYMRQID TADAAAEAID CALPQEFVSW FASNAFTVQS EALLLRYRNT LTGQLLFECK LYNEGYIALS YSGSGPLTFP TDGIFEVVSW VPRLYQLASV GSLATGRMLK Q |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Material: COPPER / Mesh: 400 | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)