+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6293 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | JRFL gp140 NFL2P unliganded | |||||||||
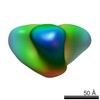
 Map data Map data | Reconstruction of JRFL gp140 NFL unliganded | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | de Val N / Sharma S / Bale S | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2015 Journal: Cell Rep / Year: 2015Title: Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Authors: Shailendra Kumar Sharma / Natalia de Val / Shridhar Bale / Javier Guenaga / Karen Tran / Yu Feng / Viktoriya Dubrovskaya / Andrew B Ward / Richard T Wyatt /  Abstract: Viral glycoproteins mediate entry by pH-activated or receptor-engaged activation and exist in metastable pre-fusogenic states that may be stabilized by directed rational design. As recently reported, ...Viral glycoproteins mediate entry by pH-activated or receptor-engaged activation and exist in metastable pre-fusogenic states that may be stabilized by directed rational design. As recently reported, the conformationally fixed HIV-1 envelope glycoprotein (Env) trimers in the pre-fusion state (SOSIP) display molecular homogeneity and structural integrity at relatively high levels of resolution. However, the SOSIPs necessitate full Env precursor cleavage, which requires endogenous furin overexpression. Here, we developed an alternative strategy using flexible peptide covalent linkage of Env subdomains to produce soluble, homogeneous, and cleavage-independent Env mimics, called native flexibly linked (NFL) trimers, as vaccine candidates. This simplified design avoids the need for furin co-expression and, in one case, antibody affinity purification to accelerate trimer scale-up for preclinical and clinical applications. We have successfully translated the NFL design to multiple HIV-1 subtypes, establishing the potential to become a general method of producing native-like, well-ordered Env trimers for HIV-1 or other viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6293.map.gz emd_6293.map.gz | 11.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6293-v30.xml emd-6293-v30.xml emd-6293.xml emd-6293.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6293.gif 400_6293.gif 80_6293.gif 80_6293.gif | 10.2 KB 1.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6293 http://ftp.pdbj.org/pub/emdb/structures/EMD-6293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6293 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6293.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6293.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of JRFL gp140 NFL unliganded | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : JRFL gp140 NFL2P unliganded
| Entire | Name: JRFL gp140 NFL2P unliganded |
|---|---|
| Components |
|
-Supramolecule #1000: JRFL gp140 NFL2P unliganded
| Supramolecule | Name: JRFL gp140 NFL2P unliganded / type: sample / ID: 1000 / Oligomeric state: trimer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 420 KDa / Theoretical: 420 KDa / Method: Size exclusion chromatography (SEC) |
-Macromolecule #1: JRFL gp140 NFL2
| Macromolecule | Name: JRFL gp140 NFL2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / synonym: HIV-1 Human immunodeficiency virus 1 / synonym: HIV-1 |
| Molecular weight | Experimental: 420 KDa / Theoretical: 420 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK 293T Homo sapiens (human) / Recombinant cell: HEK 293T |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris-HCl, 150 mM NaCl |
| Staining | Type: NEGATIVE / Details: 2% uranyl formate for 30 seconds |
| Grid | Details: 400 mesh Cu grids, negatively glow discharged for 30 seconds |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Date | May 6, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 163 / Average electron dose: 37.91 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 46000 |
| Sample stage | Specimen holder model: HOME BUILD / Tilt angle max: 50 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Software - Name: EMAN, sparx / Number images used: 16737 |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















