+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GPCR16-miniGs complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Complex / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbitter taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / ganglioside catabolic process / Class C/3 (Metabotropic glutamate/pheromone receptors) / oligosaccharide catabolic process / exo-alpha-sialidase / exo-alpha-sialidase activity / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation ...bitter taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / ganglioside catabolic process / Class C/3 (Metabotropic glutamate/pheromone receptors) / oligosaccharide catabolic process / exo-alpha-sialidase / exo-alpha-sialidase activity / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / intracellular transport / D1 dopamine receptor binding / vascular endothelial cell response to laminar fluid shear stress / activation of adenylate cyclase activity / renal water homeostasis / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / trans-Golgi network / bone development / G protein-coupled receptor activity / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / external side of plasma membrane / lysosomal membrane / GTPase activity / synapse / GTP binding / protein-containing complex binding / endoplasmic reticulum / signal transduction / extracellular exosome / extracellular region / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.84 Å | |||||||||
 Authors Authors | Wang X / Wu LJ / Hua T / Liu ZJ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2025 Journal: Cell Rep / Year: 2025Title: Structural basis of β-glucopyranoside salicin recognition by a human bitter taste GPCR. Authors: Xin Wang / Cui Zhou / Weizhen Ao / Lijie Wu / Yiran Wu / Weixiu Xu / Shenhui Liu / Qiwen Tan / Ling Wang / Fei Zhao / Junlin Liu / Yuan Pei / Suwen Zhao / Tian Hua /  Abstract: The human perception of bitterness is mediated by type 2 taste receptors (TAS2Rs), which recognize a broad array of bitter substances with distinct chemical properties. TAS2R16 exhibits a pronounced ...The human perception of bitterness is mediated by type 2 taste receptors (TAS2Rs), which recognize a broad array of bitter substances with distinct chemical properties. TAS2R16 exhibits a pronounced selectivity for β-glucoside-moiety-containing compounds, such as salicin from willow bark. However, the molecular mechanism of moiety-specific recognition and receptor activation in TAS2R16 remains unclear. Here, we present cryoelectron microscopy structures of the salicin-activated human TAS2R16 complexed with gustducin and G and G proteins. The binding mode of salicin with TAS2R16 and the specific interactions of the β-D-glucopyranoside moiety are detailed. Together with molecular docking and mutagenesis data, this study uncovers the structural underpinnings of TAS2R16's group-specific recognition, receptor activation, and subsequent gustducin and G protein coupling. These findings advance our understanding of human bitter taste receptors and provide a foundation for structural modifications of bitter glycosides, opening potential therapeutic applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62484.map.gz emd_62484.map.gz | 56.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62484-v30.xml emd-62484-v30.xml emd-62484.xml emd-62484.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_62484.png emd_62484.png | 61.8 KB | ||
| Filedesc metadata |  emd-62484.cif.gz emd-62484.cif.gz | 7.7 KB | ||
| Others |  emd_62484_additional_1.map.gz emd_62484_additional_1.map.gz emd_62484_half_map_1.map.gz emd_62484_half_map_1.map.gz emd_62484_half_map_2.map.gz emd_62484_half_map_2.map.gz | 56.9 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62484 http://ftp.pdbj.org/pub/emdb/structures/EMD-62484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62484 | HTTPS FTP |
-Related structure data
| Related structure data |  9kpdMC  9k6lC  9kpeC  9kpfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62484.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62484.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_62484_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_62484_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_62484_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GPCR16 complexed with miniGSG
| Entire | Name: GPCR16 complexed with miniGSG |
|---|---|
| Components |
|
-Supramolecule #1: GPCR16 complexed with miniGSG
| Supramolecule | Name: GPCR16 complexed with miniGSG / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms sh...
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(t) subunit alpha-3 type: protein_or_peptide / ID: 1 Details: Author stated that the map submitted was automatically generated by deepenhancer in CryoSPARC. Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.583334 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKE NLYFQSNSKT EDQRNEEKAQ REANKKIEKQ LQKDKQVYRA THRLLLLGAD NSGKSTIVKQ MRILHGGSGG SGGTSGIFE TKFQVDKVNF HMFDVGGQRD ERRKWIQCFN DVTAIIFVVD SSDYNRLQEA LNDFKSIWNN RWLRTISVIL F LNKQDLLA ...String: MDYKDDDDKE NLYFQSNSKT EDQRNEEKAQ REANKKIEKQ LQKDKQVYRA THRLLLLGAD NSGKSTIVKQ MRILHGGSGG SGGTSGIFE TKFQVDKVNF HMFDVGGQRD ERRKWIQCFN DVTAIIFVVD SSDYNRLQEA LNDFKSIWNN RWLRTISVIL F LNKQDLLA EKVLAGKSKI EDYFPEFARY TTPEDATPEP GEDPRVTRAK YFIRDEFLRI STASGDGRHY CYPHFTCAVD TQ NVKFVFD AVTDIIIKEN LKDCGLF UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.728426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWNGSSGGG GSGGGGSSGV SGWRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.845516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQVQLQESGG GLVQPGGSLR LSCAASGFTF SNYKMNWVRQ APGKGLEWVS DISQSGASIS YTGSVKGRFT ISRDNAKNTL YLQMNSLKP EDTAVYYCAR CPAPFTRDCF DVTSTTYAYR GQGTQVTVSS HHHHHH |
-Macromolecule #5: Fusion protein 1,exo-alpha-sialidase,Taste receptor type 2 member...
| Macromolecule | Name: Fusion protein 1,exo-alpha-sialidase,Taste receptor type 2 member 16,Fusion protein 2,exo-alpha-sialidase,Taste receptor type 2 member 16 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: exo-alpha-sialidase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 113.855758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAHHHHHH HHHHENLYFQ SAHHHHHHSS GLEVLFQGPP EGAALTEKTD IFESGRNGNP NKDGIKSYR IPALLKTDKG TLIAGADERR LHSSDWGDIG MVIRRSEDNG KTWGDRVTIT NLRDNPKASD PSIGSPVNID M VLVQDPET ...String: MKTIIALSYI FCLVFADYKD DDDAHHHHHH HHHHENLYFQ SAHHHHHHSS GLEVLFQGPP EGAALTEKTD IFESGRNGNP NKDGIKSYR IPALLKTDKG TLIAGADERR LHSSDWGDIG MVIRRSEDNG KTWGDRVTIT NLRDNPKASD PSIGSPVNID M VLVQDPET KRIFSIYDMF PEGKGIFGMS SQKEEAYKKI DGKTYQILYR EGEKGAYTIR ENGTVYTPDG KATDYRVVVD PV KPAYSDK GDLYKGDQLL GNIYFTTNKT SPFRIAKDSY LWMSYSDDDG KTWSAPQDIT PMVKADWMKF LGVGPGTGIV LRN GPHKGR ILIPVYTTNN VSHLDGSQSS RVIYSDDHGK TWHAGEAVND NRQVDGQKIH SSTMNNRRAQ NTESTVVQLN NGDV KLFMR GLTGDLQVAT SKDGGVTWEK DIKRYPQVKD VYVQMSAIHT MHEGKEYIIL SNAGGPKREN GMVHLARVEE NGELT WLKH NPIQKGEFAY NSLQELGNGE YGILYEHTEK GQNAYTLSFR KFNWEFLSKN GSGSGIPIQL TVFFMIIYVL ESLTII VQS SLIVAVLGRE WLQVRRLMPV DMILISLGIS RFCLQWASML NNFCSYFNLN YVLCNLTITW EFFNILTFWL NSLLTVF YC IKVSSFTHHI FLWLRWRILR LFPWILLGCL MITCVTIIPS AIGNYIQIQL LTMEHLPRNS TVTDKLENFH QYQFQAHT V ALVIPFILFL ASTIFLMASL TKQIQHHSTG HCNPSMKARF TALRSLAVLF IVFTSYFLTI LITIIGTLFD KRCWLWVWE AFVYAFILMH STSLMLSSPT LKRILKGKCG SGSGGSGSGG SGSGGSGSGS SGGVFTLEDF VGDWEQTAAY NLDQVLEQGG VSSLLQNLA VSVTPIQRIV RSGENALKID IHVIIPYEGL SADQMAQIEE VFKVVYPVDD HHFKVILPYG TLVIDGVTPN M LNYFGRPY EGIAVFDGKK ITVTGTLWNG NKIIDERLIT PDGSMLFRVT INS UniProtKB: Sialidase A, Taste receptor type 2 member 16 |
-Macromolecule #6: 2-(hydroxymethyl)phenyl beta-D-glucopyranoside
| Macromolecule | Name: 2-(hydroxymethyl)phenyl beta-D-glucopyranoside / type: ligand / ID: 6 / Number of copies: 1 / Formula: SA0 |
|---|---|
| Molecular weight | Theoretical: 286.278 Da |
| Chemical component information | 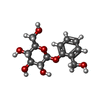 ChemComp-SA0: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-9kpd: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































