+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sarbecovirus BANAL-20-52 Spike Trimer in a Locked Conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | spike protein / VIRAL PROTEIN | |||||||||
| Biological species |  BANAL-20-52 (virus) / BANAL-20-52 (virus) /  Sarbecovirus Sarbecovirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Wang J / Xiong X | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2025 Journal: Sci Adv / Year: 2025Title: SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding. Authors: Jingjing Wang / Yong Ma / Zimu Li / Hang Yuan / Banghui Liu / Zexuan Li / Mengzhen Su / Gul Habib / Yutong Liu / Lutang Fu / Peiyi Wang / Mei Li / Jun He / Jing Chen / Peng Zhou / Zhengli ...Authors: Jingjing Wang / Yong Ma / Zimu Li / Hang Yuan / Banghui Liu / Zexuan Li / Mengzhen Su / Gul Habib / Yutong Liu / Lutang Fu / Peiyi Wang / Mei Li / Jun He / Jing Chen / Peng Zhou / Zhengli Shi / Xinwen Chen / Xiaoli Xiong /  Abstract: High-affinity and specific binding toward the human angiotensin-converting enzyme 2 (hACE2) receptor by severe acute respiratory syndrome coronavirus (SARS)-related coronaviruses (SARSr-CoVs) remains ...High-affinity and specific binding toward the human angiotensin-converting enzyme 2 (hACE2) receptor by severe acute respiratory syndrome coronavirus (SARS)-related coronaviruses (SARSr-CoVs) remains incompletely understood. We report cryo-electron microscopy structures of eight different S-proteins from SARSr-CoVs found across Asia, Europe, and Africa. These S-proteins all adopt tightly packed, locked, prefusion conformations. These structures enable the classification of SARSr-CoV S-proteins into three types, based on their receptor-binding motif (RBM) structures and ACE2 binding characteristics. Type-2 S-proteins often preferentially bind bat ACE2 (bACE2) over hACE2. We report a structure of a type-2 BtKY72-RBD in complex with bACE2 to understand ACE2 specificity. Structure-guided mutagenesis of BtKY72-RBD reveals that multiple synergistic mutations in four different regions of RBM are required to achieve high-affinity hACE2 binding. Similar RBM changes can also confer hACE2 binding to another type-2 BM48-31 S-protein, which is primarily non-ACE2 binding. These results provide an understanding of how high-affinity hACE2 binding may be acquired by SARSr-CoV S-proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60553.map.gz emd_60553.map.gz | 89.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60553-v30.xml emd-60553-v30.xml emd-60553.xml emd-60553.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60553.png emd_60553.png | 73.1 KB | ||
| Filedesc metadata |  emd-60553.cif.gz emd-60553.cif.gz | 7.3 KB | ||
| Others |  emd_60553_half_map_1.map.gz emd_60553_half_map_1.map.gz emd_60553_half_map_2.map.gz emd_60553_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60553 http://ftp.pdbj.org/pub/emdb/structures/EMD-60553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60553 | HTTPS FTP |
-Validation report
| Summary document |  emd_60553_validation.pdf.gz emd_60553_validation.pdf.gz | 821.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60553_full_validation.pdf.gz emd_60553_full_validation.pdf.gz | 820.8 KB | Display | |
| Data in XML |  emd_60553_validation.xml.gz emd_60553_validation.xml.gz | 15.2 KB | Display | |
| Data in CIF |  emd_60553_validation.cif.gz emd_60553_validation.cif.gz | 18.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60553 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60553 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60553 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60553 | HTTPS FTP |
-Related structure data
| Related structure data |  8zy2MC  8zy0C  8zy1C  8zy3C  8zy4C  8zy5C  8zy6C  8zy7C  8zy9C  8zyaC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60553.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60553.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.168 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60553_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
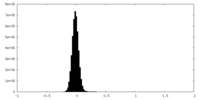
| Density Histograms |
-Half map: #1
| File | emd_60553_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
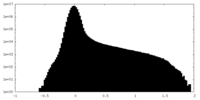
| Density Histograms |
- Sample components
Sample components
-Entire : Bat coronavirus BANAL-20-52
| Entire | Name: Bat coronavirus BANAL-20-52 |
|---|---|
| Components |
|
-Supramolecule #1: Bat coronavirus BANAL-20-52
| Supramolecule | Name: Bat coronavirus BANAL-20-52 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: the spike trimer |
|---|---|
| Source (natural) | Organism:  BANAL-20-52 (virus) BANAL-20-52 (virus) |
| Molecular weight | Theoretical: 420 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sarbecovirus Sarbecovirus |
| Molecular weight | Theoretical: 125.744578 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SSTRGVYYPD KVFRSSVLHL TQDLFLPFFS NVTWFHAIHV SGTNGIKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHRNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SSTRGVYYPD KVFRSSVLHL TQDLFLPFFS NVTWFHAIHV SGTNGIKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHRNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPP GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCSLDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATT FASVYAWNRK RISNCVADYS VLYNSTSFST FKCYGVSPTK LNDLCFTNVY ADSF VVRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFHPTNGVG YQPYRVVVLS FELLNAPATV CGPKKSTNLI KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN ASNQVAVLYQ DVNCTEVPVA IHANQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSRS VASQSIIAYT MSLGAENSVA YSNNSIAIPT NFTISVTT E ILPVSMTKTS VDCTMYICGD STECSNLLLQ YGSFCTQLNR ALTGIAVEQD KNTQEVFAQV KQIYKTPQIK DFGGFNFSQ ILPDPSKPSK RSFIEDLLFN KVTLADAGFI KQYGDCLGDI AARDLICAQK FNGLTVLPPL LTDEMIAQYT SALLAGTITS GWTFGAGAA LQIPFAMQMA YRFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLSSTAS ALGKLQDVVN QNAQALNTLV K QLSSNFGA ISSVLNDILS RLDKVEAEVQ IDRLITGRLQ SLQTYVTQQL IRAAEIRASA NLAATKMSEC VLGQSKRVDF CG KGYHLMS FPQSAPHGVV FLHVTYVPAQ EKNFTTAPAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSG NCDVVI GIVNNTVYDP |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 33 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: BILIVERDINE IX ALPHA
| Macromolecule | Name: BILIVERDINE IX ALPHA / type: ligand / ID: 4 / Number of copies: 3 / Formula: BLA |
|---|---|
| Molecular weight | Theoretical: 582.646 Da |
| Chemical component information |  ChemComp-BLA: |
-Macromolecule #5: LINOLEIC ACID
| Macromolecule | Name: LINOLEIC ACID / type: ligand / ID: 5 / Number of copies: 3 / Formula: EIC |
|---|---|
| Molecular weight | Theoretical: 280.445 Da |
| Chemical component information |  ChemComp-EIC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4. | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)