[English] 日本語
 Yorodumi
Yorodumi- EMDB-5957: Single particle EM reveals plasticity of interactions between the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5957 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle EM reveals plasticity of interactions between the adenovirus penton base and integrin alphaV-beta3 | |||||||||
 Map data Map data | Reconstruction of unliganded integrin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adenovirus / integrin / virus infection / electron microscopy / fluorescence correlation microscopy | |||||||||
| Function / homology |  Function and homology information Function and homology informationintegrin alphav-beta8 complex / integrin alphav-beta6 complex / transforming growth factor beta production / negative regulation of entry of bacterium into host cell / integrin alphav-beta5 complex / opsonin binding / integrin alphav-beta1 complex / Cross-presentation of particulate exogenous antigens (phagosomes) / extracellular matrix protein binding / Laminin interactions ...integrin alphav-beta8 complex / integrin alphav-beta6 complex / transforming growth factor beta production / negative regulation of entry of bacterium into host cell / integrin alphav-beta5 complex / opsonin binding / integrin alphav-beta1 complex / Cross-presentation of particulate exogenous antigens (phagosomes) / extracellular matrix protein binding / Laminin interactions / integrin alphav-beta3 complex / negative regulation of lipoprotein metabolic process / entry into host cell by a symbiont-containing vacuole / alphav-beta3 integrin-PKCalpha complex / alphav-beta3 integrin-HMGB1 complex / negative regulation of lipid transport / regulation of phagocytosis / Elastic fibre formation / alphav-beta3 integrin-IGF-1-IGF1R complex / transforming growth factor beta binding / positive regulation of small GTPase mediated signal transduction / filopodium membrane / extracellular matrix binding / wound healing, spreading of epidermal cells / apolipoprotein A-I-mediated signaling pathway / negative regulation of low-density lipoprotein particle clearance / apoptotic cell clearance / integrin complex / heterotypic cell-cell adhesion / Molecules associated with elastic fibres / cell adhesion mediated by integrin / negative chemotaxis / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / Syndecan interactions / positive regulation of osteoblast proliferation / microvillus membrane / cell-substrate adhesion / endodermal cell differentiation / PECAM1 interactions / TGF-beta receptor signaling activates SMADs / fibronectin binding / lamellipodium membrane / positive regulation of intracellular signal transduction / negative regulation of macrophage derived foam cell differentiation / negative regulation of lipid storage / ECM proteoglycans / Integrin cell surface interactions / vasculogenesis / specific granule membrane / voltage-gated calcium channel activity / coreceptor activity / phagocytic vesicle / ERK1 and ERK2 cascade / extrinsic apoptotic signaling pathway in absence of ligand / substrate adhesion-dependent cell spreading / positive regulation of cell adhesion / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / protein kinase C binding / cell-matrix adhesion / integrin-mediated signaling pathway / Signal transduction by L1 / negative regulation of extrinsic apoptotic signaling pathway / cell-cell adhesion / VEGFA-VEGFR2 Pathway / calcium ion transmembrane transport / integrin binding / ruffle membrane / cell migration / positive regulation of cytosolic calcium ion concentration / virus receptor activity / protease binding / angiogenesis / cell adhesion / positive regulation of cell migration / external side of plasma membrane / focal adhesion / positive regulation of cell population proliferation / Neutrophil degranulation / symbiont entry into host cell / cell surface / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 44.0 Å | |||||||||
 Authors Authors | Veesler D / Cupelli K / Burger M / Graber P / Stehle T / Johnson JE | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2014 Journal: Proc Natl Acad Sci U S A / Year: 2014Title: Single-particle EM reveals plasticity of interactions between the adenovirus penton base and integrin αVβ3. Authors: David Veesler / Karolina Cupelli / Markus Burger / Peter Gräber / Thilo Stehle / John E Johnson /   Abstract: Human adenoviruses are double-stranded DNA viruses responsible for numerous infections, some of which can be fatal. Furthermore, adenoviruses are currently used in clinical trials as vectors for gene ...Human adenoviruses are double-stranded DNA viruses responsible for numerous infections, some of which can be fatal. Furthermore, adenoviruses are currently used in clinical trials as vectors for gene therapy applications. Although initial binding of adenoviruses to host attachment receptors has been extensively characterized, the interactions with the entry receptor (integrins) remain poorly understood at the structural level. We characterized the interactions between the adenovirus 9 penton base subunit and αVβ3 integrin using fluorescence correlation spectroscopy and single-particle electron microscopy to understand the mechanisms underlying virus internalization and infection. Our results indicate that the penton base subunit can bind integrins with high affinity and in several different orientations. These outcomes correlate with the requirement of the pentameric penton base to simultaneously bind several integrins to enable their clustering and promote virus entry into the host cell. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5957.map.gz emd_5957.map.gz | 2.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5957-v30.xml emd-5957-v30.xml emd-5957.xml emd-5957.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| Images |  400_5957.gif 400_5957.gif 80_5957.gif 80_5957.gif | 20.5 KB 2.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5957 http://ftp.pdbj.org/pub/emdb/structures/EMD-5957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5957 | HTTPS FTP |
-Related structure data
| Related structure data |  5955C  5956C  5958C  5959C  5960C  5961C  5962C  5963C  5964C  5965C  5966C  5967C  5968C  5969C  5970C  5971C  5972C  5973C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5957.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5957.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of unliganded integrin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
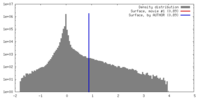
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human alphaV-beta3 integrin ectodomain
| Entire | Name: Human alphaV-beta3 integrin ectodomain |
|---|---|
| Components |
|
-Supramolecule #1000: Human alphaV-beta3 integrin ectodomain
| Supramolecule | Name: Human alphaV-beta3 integrin ectodomain / type: sample / ID: 1000 / Oligomeric state: heterodimer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: alphaV-beta3 integrin ectodomain
| Macromolecule | Name: alphaV-beta3 integrin ectodomain / type: protein_or_peptide / ID: 1 / Details: Second UniProt identifier: P05106 / Number of copies: 1 / Oligomeric state: heterodimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 180 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Integrin alpha-V |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 10 mM Tris, 150 mM NaCl, 2 mM MnCl2 |
|---|---|
| Staining | Type: NEGATIVE / Details: 2% uranyl formate |
| Grid | Details: C-flat 2/0.5 grids overlaid with a thin layer of carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Jun 13, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 540 / Average electron dose: 33 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.55 µm / Nominal defocus min: 0.1 µm / Nominal magnification: 85714 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Random conical tilt reconstruction |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 44.0 Å / Resolution method: OTHER / Software - Name: Spider / Number images used: 769 |
| Final two d classification | Number classes: 1 |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)