+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5419 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sulfolobus solfataricus methylation-guide sRNP | |||||||||
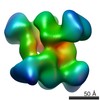
 Map data Map data | Projection matching single particle reconstruction of Sulfolobus solfataricus methylation-guide sRNP, 26 Angstrom resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | archaea / box C/D sRNP / single particle EM / rRNA modification / RNA-protein complex / non-coding RNA | |||||||||
| Biological species |   Sulfolobus solfataricus (archaea) Sulfolobus solfataricus (archaea) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Bower-Phipps KR / Taylor DW / Wang HW / Baserga SJ | |||||||||
 Citation Citation |  Journal: RNA / Year: 2012 Journal: RNA / Year: 2012Title: The box C/D sRNP dimeric architecture is conserved across domain Archaea. Authors: Kathleen R Bower-Phipps / David W Taylor / Hong-Wei Wang / Susan J Baserga /  Abstract: Box C/D small (nucleolar) ribonucleoproteins [s(no)RNPs] catalyze RNA-guided 2'-O-ribose methylation in two of the three domains of life. Recent structural studies have led to a controversy over ...Box C/D small (nucleolar) ribonucleoproteins [s(no)RNPs] catalyze RNA-guided 2'-O-ribose methylation in two of the three domains of life. Recent structural studies have led to a controversy over whether box C/D sRNPs functionally assemble as monomeric or dimeric macromolecules. The archaeal box C/D sRNP from Methanococcus jannaschii (Mj) has been shown by glycerol gradient sedimentation, gel filtration chromatography, native gel analysis, and single-particle electron microscopy (EM) to adopt a di-sRNP architecture, containing four copies of each box C/D core protein and two copies of the Mj sR8 sRNA. Subsequently, investigators used a two-stranded artificial guide sRNA, CD45, to assemble a box C/D sRNP from Sulfolobus solfataricus with a short RNA methylation substrate, yielding a crystal structure of a mono-sRNP. To more closely examine box C/D sRNP architecture, we investigate the role of the omnipresent sRNA loop as a structural determinant of sRNP assembly. We show through sRNA mutagenesis, native gel electrophoresis, and single-particle EM that a di-sRNP is the near exclusive architecture obtained when reconstituting box C/D sRNPs with natural or artificial sRNAs containing an internal loop. Our results span three distantly related archaeal species--Sulfolobus solfataricus, Pyrococcus abyssi, and Archaeoglobus fulgidus--indicating that the di-sRNP architecture is broadly conserved across the entire archaeal domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5419.map.gz emd_5419.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5419-v30.xml emd-5419-v30.xml emd-5419.xml emd-5419.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5419_1.png emd_5419_1.png | 917 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5419 http://ftp.pdbj.org/pub/emdb/structures/EMD-5419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5419 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5419.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5419.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Projection matching single particle reconstruction of Sulfolobus solfataricus methylation-guide sRNP, 26 Angstrom resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Sulfolobus solfataricus methylation-guide sRNP
| Entire | Name: Sulfolobus solfataricus methylation-guide sRNP |
|---|---|
| Components |
|
-Supramolecule #1000: Sulfolobus solfataricus methylation-guide sRNP
| Supramolecule | Name: Sulfolobus solfataricus methylation-guide sRNP / type: sample / ID: 1000 Oligomeric state: Four copies of each box C/D core protein could be docked in this map. Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 371 KDa |
-Macromolecule #1: ribonucleoprotein
| Macromolecule | Name: ribonucleoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: box C/D sRNP / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Sulfolobus solfataricus (archaea) Sulfolobus solfataricus (archaea) |
| Molecular weight | Theoretical: 371 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 20mM HEPES, 500mM NaCl, 1.5mM MgCl2 |
|---|---|
| Staining | Type: NEGATIVE / Details: 2% uranyl formate |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Details | low-dose conditions |
| Date | Jun 1, 2011 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 6149 |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, Situs |
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















