[English] 日本語
 Yorodumi
Yorodumi- EMDB-53283: Single particle cryo-EM structure of MdtF V610F with bound Rhodam... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo-EM structure of MdtF V610F with bound Rhodamine 6G | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | multidrug / pump / anaerobic / lipid / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationxenobiotic transmembrane transport / bile acid transmembrane transporter activity / bile acid and bile salt transport / efflux transmembrane transporter activity / xenobiotic transmembrane transporter activity / xenobiotic transport / response to toxic substance / response to antibiotic / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Lawrence R / Adams C / Sousa JS / Ahdash Z / Reading E | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2025 Journal: bioRxiv / Year: 2025Title: Molecular basis for multidrug efflux by an anaerobic RND transporter. Authors: Ryan Lawrence / Mohd Athar / Muhammad R Uddin / Christopher Adams / Joana S Sousa / Oliver Durrant / Sophie Lellman / Lucy Sutton / C William Keevil / Nisha Patel / Christine Prosser / David ...Authors: Ryan Lawrence / Mohd Athar / Muhammad R Uddin / Christopher Adams / Joana S Sousa / Oliver Durrant / Sophie Lellman / Lucy Sutton / C William Keevil / Nisha Patel / Christine Prosser / David McMillan / Helen I Zgurskaya / Attilio V Vargiu / Zainab Ahdash / Eamonn Reading Abstract: Bacteria can resist antibiotics and toxic substances within demanding ecological settings, such as low oxygen, extreme acid, and during nutrient starvation. MdtEF, a proton motive force-driven efflux ...Bacteria can resist antibiotics and toxic substances within demanding ecological settings, such as low oxygen, extreme acid, and during nutrient starvation. MdtEF, a proton motive force-driven efflux pump from the resistance-nodulation-cell division (RND) superfamily, is upregulated in these conditions but its molecular mechanism is unknown. Here, we report cryo-electron microscopy structures of Escherichia coli multidrug transporter MdtF within native-lipid nanodiscs, including a single-point mutant with an altered multidrug phenotype and associated substrate-bound form. We reveal that drug binding domain and channel conformational plasticity likely governs promiscuous substrate specificity, analogous to its closely related, constitutively expressed counterpart, AcrB. Whereas we discover distinct transmembrane state transitions within MdtF, which create a more engaged proton relay network, altered drug transport allostery and an acid-responsive increase in efflux efficiency. Physiologically, this provides means of xenobiotic and metabolite disposal within remodelled cell membranes that presage encounters with acid stresses, as endured in the gastrointestinal tract. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_53283.map.gz emd_53283.map.gz | 62.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-53283-v30.xml emd-53283-v30.xml emd-53283.xml emd-53283.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_53283_fsc.xml emd_53283_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_53283.png emd_53283.png | 94.5 KB | ||
| Filedesc metadata |  emd-53283.cif.gz emd-53283.cif.gz | 6.8 KB | ||
| Others |  emd_53283_half_map_1.map.gz emd_53283_half_map_1.map.gz emd_53283_half_map_2.map.gz emd_53283_half_map_2.map.gz | 51.9 MB 51.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-53283 http://ftp.pdbj.org/pub/emdb/structures/EMD-53283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53283 | HTTPS FTP |
-Related structure data
| Related structure data |  9qptMC  9qprC  9qpsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_53283.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_53283.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.947 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_53283_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_53283_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : R6G-bound homotrimer of V610F multidrug resistance protein MdtF i...
| Entire | Name: R6G-bound homotrimer of V610F multidrug resistance protein MdtF in SMALP nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: R6G-bound homotrimer of V610F multidrug resistance protein MdtF i...
| Supramolecule | Name: R6G-bound homotrimer of V610F multidrug resistance protein MdtF in SMALP nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 334.551 KDa |
-Macromolecule #1: Multidrug resistance protein MdtF
| Macromolecule | Name: Multidrug resistance protein MdtF / type: protein_or_peptide / ID: 1 Details: V610F MdtF protein with a C-terminal 6x His (hexahistidine) tag, separated by a linker sequence including a TEV cleavage site Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 113.807898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANYFIDRPV FAWVLAIIMM LAGGLAIMNL PVAQYPQIAP PTITVSATYP GADAQTVEDS VTQVIEQNMN GLDGLMYMSS TSDAAGNAS ITLTFETGTS PDIAQVQVQN KLQLAMPSLP EAVQQQGISV DKSSSNILMV AAFISDNGSL NQYDIADYVA S NIKDPLSR ...String: MANYFIDRPV FAWVLAIIMM LAGGLAIMNL PVAQYPQIAP PTITVSATYP GADAQTVEDS VTQVIEQNMN GLDGLMYMSS TSDAAGNAS ITLTFETGTS PDIAQVQVQN KLQLAMPSLP EAVQQQGISV DKSSSNILMV AAFISDNGSL NQYDIADYVA S NIKDPLSR TAGVGSVQLF GSEYAMRIWL DPQKLNKYNL VPSDVISQIK VQNNQISGGQ LGGMPQAADQ QLNASIIVQT RL QTPEEFG KILLKVQQDG SQVLLRDVAR VELGAEDYST VARYNGKPAA GIAIKLAAGA NALDTSRAVK EELNRLSAYF PAS LKTVYP YDTTPFIEIS IQEVFKTLVE AIILVFLVMY LFLQNFRATI IPTIAVPVVI LGTFAILSAV GFTINTLTMF GMVL AIGLL VDDAIVVVEN VERVIAEDKL PPKEATHKSM GQIQRALVGI AVVLSAVFMP MAFMSGATGE IYRQFSITLI SSMLL SVFV AMSLTPALCA TILKAAPEGG HKPNALFARF NTLFEKSTQH YTDSTRSLLR CTGRYMVVYL LICAGMAVLF LRTPTS FLP EEDQGVFMTT AQLPSGATMV NTTKVLQQVT DYYLTKEKDN VQSVFTFGGF GFSGQGQNNG LAFISLKPWS ERVGEEN SV TAIIQRAMIA LSSINKAVVF PFNLPAVAEL GTASGFDMEL LDNGNLGHEK LTQARNELLS LAAQSPNQVT GVRPNGLE D TPMFKVNVNA AKAEAMGVAL SDINQTISTA FGSSYVNDFL NQGRVKKVYV QAGTPFRMLP DNINQWYVRN ASGTMAPLS AYSSTEWTYG SPRLERYNGI PSMEILGEAA AGKSTGDAMK FMADLVAKLP AGVGYSWTGL SYQEALSSNQ APALYAISLV VVFLALAAL YESWSIPFSV MLVVPLGVVG ALLATDLRGL SNDVYFQVGL LTTIGLSAKN AILIVEFAVE MMQKEGKTPI E AIIEAARM RLRPILMTSL AFILGVLPLV ISHGAGSGAQ NAVGTGVMGG MFAATVLAIY FVPVFFVVVE HLFARFKKAA SG ENLYFQS LEHHHHHH UniProtKB: Multidrug resistance protein MdtF |
-Macromolecule #2: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 2 / Number of copies: 12 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Macromolecule #3: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 3 / Number of copies: 23 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #4: RHODAMINE 6G
| Macromolecule | Name: RHODAMINE 6G / type: ligand / ID: 4 / Number of copies: 1 / Formula: RHQ |
|---|---|
| Molecular weight | Theoretical: 443.557 Da |
| Chemical component information | 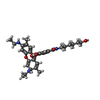 ChemComp-R6G: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 41.25 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 150000 |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)