[English] 日本語
 Yorodumi
Yorodumi- EMDB-51902: Cryo-EM structure of lipid-bound human myoferlin (25 mol% DOPS/5 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of lipid-bound human myoferlin (25 mol% DOPS/5 mol% PI(4,5)P2 nanodisc) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ferlin / multi-C2 domain protein / tail-anchored membrane protein / vesicle docking and fusion / membrane remodeling / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of neurotransmitter secretion / plasma membrane repair / blood circulation / ciliary tip / muscle contraction / caveola / phospholipid binding / centriolar satellite / nuclear envelope / synaptic vesicle membrane ...regulation of neurotransmitter secretion / plasma membrane repair / blood circulation / ciliary tip / muscle contraction / caveola / phospholipid binding / centriolar satellite / nuclear envelope / synaptic vesicle membrane / late endosome membrane / cytoplasmic vesicle / nuclear membrane / cilium / ciliary basal body / calcium ion binding / extracellular exosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.56 Å | |||||||||
 Authors Authors | Cretu C / Moser T / Preobraschenski J | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2025 Journal: EMBO J / Year: 2025Title: Structural insights into lipid membrane binding by human ferlins. Authors: Constantin Cretu / Aleksandar Chernev / Csaba Zoltán Kibédi Szabó / Vladimir Pena / Henning Urlaub / Tobias Moser / Julia Preobraschenski /   Abstract: Ferlins are ancient membrane proteins with a unique architecture, and play central roles in crucial processes that involve Ca-dependent vesicle fusion. Despite their links to multiple human diseases ...Ferlins are ancient membrane proteins with a unique architecture, and play central roles in crucial processes that involve Ca-dependent vesicle fusion. Despite their links to multiple human diseases and numerous functional studies, a mechanistic understanding of how these multi-C domain-containing proteins interact with lipid membranes to promote membrane remodelling and fusion is currently lacking. Here we obtain near-complete cryo-electron microscopy structures of human myoferlin and dysferlin in their Ca- and lipid-bound states. We show that ferlins adopt compact, ring-like tertiary structures upon membrane binding. The top arch of the ferlin ring, composed of the CC-CD region, is rigid and exhibits only little variability across the observed functional states. In contrast, the N-terminal CB and the C-terminal CF-CG domains cycle between alternative conformations and, in response to Ca, close the ferlin ring, promoting tight interaction with the target membrane. Probing key domain interfaces validates the observed architecture, and informs a model of how ferlins engage lipid bilayers in a Ca-dependent manner. This work reveals the general principles of human ferlin structures and provides a framework for future analyses of ferlin-dependent cellular functions and disease mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51902.map.gz emd_51902.map.gz | 88.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51902-v30.xml emd-51902-v30.xml emd-51902.xml emd-51902.xml | 26 KB 26 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51902_fsc.xml emd_51902_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_51902.png emd_51902.png | 33.4 KB | ||
| Masks |  emd_51902_msk_1.map emd_51902_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-51902.cif.gz emd-51902.cif.gz | 8.4 KB | ||
| Others |  emd_51902_additional_1.map.gz emd_51902_additional_1.map.gz emd_51902_half_map_1.map.gz emd_51902_half_map_1.map.gz emd_51902_half_map_2.map.gz emd_51902_half_map_2.map.gz | 87.9 MB 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51902 http://ftp.pdbj.org/pub/emdb/structures/EMD-51902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51902 | HTTPS FTP |
-Related structure data
| Related structure data |  9h6xMC  9qkvC  9qleC  9qlfC  9qlnC  9qlsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51902.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51902.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.72 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_51902_msk_1.map emd_51902_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_51902_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_51902_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_51902_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Soluble human myoferlin (residues 1-1997) bound to an MSP2N2-base...
| Entire | Name: Soluble human myoferlin (residues 1-1997) bound to an MSP2N2-based lipid nanodisc (comprising 25 mol% DOPS and 5 mol% PI(4,5)P2) |
|---|---|
| Components |
|
-Supramolecule #1: Soluble human myoferlin (residues 1-1997) bound to an MSP2N2-base...
| Supramolecule | Name: Soluble human myoferlin (residues 1-1997) bound to an MSP2N2-based lipid nanodisc (comprising 25 mol% DOPS and 5 mol% PI(4,5)P2) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Myoferlin
| Macromolecule | Name: Myoferlin / type: protein_or_peptide / ID: 1 / Details: Fer-1-like protein 3, FER1L3, MYOF / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 232.625953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASWSHPQFE KGGGSGGGSG GGSWSHPQFE KLEVLFQGPG SGDKDCEQSN AMLRVIVESA SNIPKTKFGK PDPIVSVIFK DEKKKTKKV DNELNPVWNE ILEFDLRGIP LDFSSSLGII VKDFETIGQN KLIGTATVAL KDLTGDQSRS LPYKLISLLN E RGQDTGAT ...String: MASWSHPQFE KGGGSGGGSG GGSWSHPQFE KLEVLFQGPG SGDKDCEQSN AMLRVIVESA SNIPKTKFGK PDPIVSVIFK DEKKKTKKV DNELNPVWNE ILEFDLRGIP LDFSSSLGII VKDFETIGQN KLIGTATVAL KDLTGDQSRS LPYKLISLLN E RGQDTGAT IDLVIGYDPP SAPHPNDLSG PSVPGMGGDG EEDEGDEDRL DNAVRGPGPK GPVGTVSEAQ LARRLTKVKN SR RMLSNKP QDFQIRVRVI EGRQLSGNNI RPVVKVHVCG QTHRTRIKRG NNPFFDELFF YNVNMTPSEL MDEIISIRVY NSH SLRADC LMGEFKIDVG FVYDEPGHAV MRKWLLLNDP EDTSSGSKGY MKVSMFVLGT GDEPPPERRD RDNDSDDVES NLLL PAGIA LRWVTFLLKI YRAEDIPQMD DAFSQTVKEI FGGNADKKNL VDPFVEVSFA GKKVCTNIIE KNANPEWNQV VNLQI KFPS VCEKIKLTIY DWDRLTKNDV VGTTYLHLSK IAASGGEVED FSSSGTGAAS YTVNTGETEV GFVPTFGPCY LNLYGS PRE YTGFPDPYDE LNTGKGEGVA YRGRILVELA TFLEKTPPDK KLEPISNDDL LVVEKYQRRR KYSLSAVFHS ATMLQDV GE AIQFEVSIGN YGNRFDTTCK PLASTTQYSR AVFDGNYYYY LPWAHTKPVV TLTSYWEDIS HRLDAVNTLL AMAERLQT N IEALKSGIQG KIPANQLAEL WLKLIDEVIE DTRYTLPLTE GKANVTVLDT QIRKLRSRSL SQIHEAAVRM RSEATDVKS TLAEIEDWLD KLMQLTEEPQ NSMPDIIIWM IRGEKRLAYA RIPAHQVLYS TSGENASGKY CGKTQTIFLK YPQEKNNGPK VPVELRVNI WLGLSAVEKK FNSFAEGTFT VFAEMYENQA LMFGKWGTSG LVGRHKFSDV TGKIKLKREF FLPPKGWEWE G EWIVDPER SLLTEADAGH TEFTDEVYQN ESRYPGGDWK PAEDTYTDAN GDKAASPSEL TCPPGWEWED DAWSYDINRA VD EKGWEYG ITIPPDHKPK SWVAAEKMYH THRRRRLVRK RKKDLTQTAS STARAMEELQ DQEGWEYASL IGWKFHWKQR SSD TFRRRR WRRKMAPSET HGAAAIFKLE GALGADTTED GDEKSLEKQK HSATTVFGAN TPIVSCNFDR VYIYHLRCYV YQAR NLLAL DKDSFSDPYA HICFLHRSKT TEIIHSTLNP TWDQTIIFDE VEIYGEPQTV LQNPPKVIME LFDNDQVGKD EFLGR SIFS PVVKLNSEMD ITPKLLWHPV MNGDKACGDV LVTAELILRG KDGSNLPILP PQRAPNLYMV PQGIRPVVQL TAIEIL AWG LRNMKNFQMA SITSPSLVVE CGGERVESVV IKNLKKTPNF PSSVLFMKVF LPKEELYMPP LVIKVIDHRQ FGRKPVV GQ CTIERLDRFR CDPYAGKEDI VPQLKASLLS APPCRDIVIE MEDTKPLLAS KLTEKEEEIV DWWSKFYASS GEHEKCGQ Y IQKGYSKLKI YNCELENVAE FEGLTDFSDT FKLYRGKSDE NEDPSVVGEF KGSFRIYPLP DDPSVPAPPR QFRELPDSV PQECTVRIYI VRGLELQPQD NNGLCDPYIK ITLGKKVIED RDHYIPNTLN PVFGRMYELS CYLPQEKDLK ISVYDYDTFT RDEKVGETI IDLENRFLSR FGSHCGIPEE YCVSGVNTWR DQLRPTQLLQ NVARFKGFPQ PILSEDGSRI RYGGRDYSLD E FEANKILH QHLGAPEERL ALHILRTQGL VPEHVETRTL HSTFQPNISQ GKLQMWVDVF PKSLGPPGPP FNITPRKAKK YY LRVIIWN TKDVILDEKS ITGEEMSDIY VKGWVPGNEE NKQKTDVHYR SLDGEGNFNW RFVFPFDYLP AEQLCIVAKK EHF WSIDQT EFRIPPRLII QIWDNDKFSL DDYLGFLELD LRHTIIPAKS PEKCRLDMIP DLKAMNPLKA KTASLFEQKS MKGW WPCYA EKDGARVMAG KVEMTLEILN EKEADERPAG KGRDEPNMNP KLD UniProtKB: Myoferlin |
-Macromolecule #2: 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE
| Macromolecule | Name: 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE / type: ligand / ID: 2 / Number of copies: 2 / Formula: PSF |
|---|---|
| Molecular weight | Theoretical: 455.437 Da |
| Chemical component information | 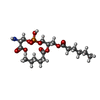 ChemComp-PSF: |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 10 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: Plasma Cleaner model PDC-32G-2 (Harrick Plasma) | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | The myoferlin-nanodisc complex was assembled in vitro, crosslinked with 0.07% (v/v) glutaraldehyde, and purified by size-exclusion chromatography on Superose 6 10/300 prior to cryo-EM grid preparation |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Software | Name: EPU (ver. v3.6.0) |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 31039 / Average exposure time: 3.16 sec. / Average electron dose: 39.595 e/Å2 Details: dataset 1: 8660 movies, 39.595 e-/A2 dataset 2: 22379 movies, 39.216 e-/A2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Software | Name: UCSF ChimeraX (ver. v1.8) |
| Details | Iterative model building and refinement with ISOLDE, Coot, and Phenix (phenix.real_space_refine) |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 152.37 |
| Output model |  PDB-9h6x: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































