[English] 日本語
 Yorodumi
Yorodumi- EMDB-50930: Rhinovirus A2 uncoating intermediate revealing the natural pocket... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rhinovirus A2 uncoating intermediate revealing the natural pocket factor (pH 5.8 and 4 degrees Celsius) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Uncoating Intermediate / Pocket Factor / Lauric Acid / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  rhinovirus A2 rhinovirus A2 | |||||||||
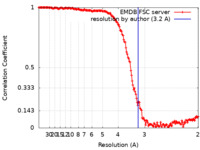
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Real-Hohn A / Blaas D | |||||||||
| Funding support |  Austria, 1 items Austria, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2025 Journal: Sci Rep / Year: 2025Title: New rhinovirus uncoating intermediate reveals how sodium versus potassium ions influence RNA release. Authors: Antonio Real-Hohn / Dieter Blaas /  Abstract: Electron microscopy (EM) of rhinovirus A2 (RV-A2) incubated in Na phosphate buffer (pH 7.6) for 12 h at 25 °C revealed partial fragmentation, whereas upon incubation in K phosphate buffer, RV-A2 ...Electron microscopy (EM) of rhinovirus A2 (RV-A2) incubated in Na phosphate buffer (pH 7.6) for 12 h at 25 °C revealed partial fragmentation, whereas upon incubation in K phosphate buffer, RV-A2 appeared intact. In buffers adjusted to pH 5.8, these differences became more pronounced; acidic Na phosphate buffer promoted disintegration of the particles, whereas in acidic K phosphate buffer, the virus appeared like native. Incubation in the acidic buffers for one hour at 4 °C followed by neutralisation resulted in the respective formation of non-infectious A particles (in Na) and a non-infectious novel uncoating intermediate (in K), which we termed 'E0 particle'. Negative staining EM revealed phosphotungstate penetration into A particles, but not into E0 particles. Cryo-EM image reconstruction of the E0 particle showed clear differences between A and E0 particles; like native virus, E0 contained VP4 and a pocket factor. Native RV-A2 RNA cores, obtained by gentle proteinase-K digestion in K and Na phosphate buffer, respectively, differed in accessibility of dsRNA regions, detected by PaSTRy. Variance in RNA compactness observed in K versus Na phosphate buffer was confirmed by rotary shadowing EM; in K phosphate buffer, the RNA remained condensed while, in Na phosphate buffer, distinct unfolding stages were apparent. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50930.map.gz emd_50930.map.gz | 88.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50930-v30.xml emd-50930-v30.xml emd-50930.xml emd-50930.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50930_fsc.xml emd_50930_fsc.xml | 20.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_50930.png emd_50930.png | 127.7 KB | ||
| Filedesc metadata |  emd-50930.cif.gz emd-50930.cif.gz | 6.8 KB | ||
| Others |  emd_50930_half_map_1.map.gz emd_50930_half_map_1.map.gz emd_50930_half_map_2.map.gz emd_50930_half_map_2.map.gz | 273.6 MB 273.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50930 http://ftp.pdbj.org/pub/emdb/structures/EMD-50930 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50930 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50930 | HTTPS FTP |
-Validation report
| Summary document |  emd_50930_validation.pdf.gz emd_50930_validation.pdf.gz | 1000.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50930_full_validation.pdf.gz emd_50930_full_validation.pdf.gz | 999.9 KB | Display | |
| Data in XML |  emd_50930_validation.xml.gz emd_50930_validation.xml.gz | 23.4 KB | Display | |
| Data in CIF |  emd_50930_validation.cif.gz emd_50930_validation.cif.gz | 30.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50930 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50930 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50930 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50930 | HTTPS FTP |
-Related structure data
| Related structure data |  9g0bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50930.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50930.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.998 Å | ||||||||||||||||||||||||||||||||||||
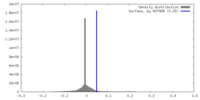
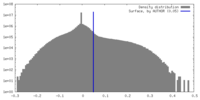
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_50930_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
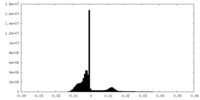
| Density Histograms |
-Half map: #1
| File | emd_50930_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
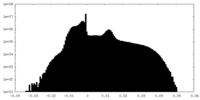
| Density Histograms |
- Sample components
Sample components
-Entire : rhinovirus A2
| Entire | Name:  rhinovirus A2 rhinovirus A2 |
|---|---|
| Components |
|
-Supramolecule #1: rhinovirus A2
| Supramolecule | Name: rhinovirus A2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / NCBI-ID: 12130 / Sci species name: rhinovirus A2 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Diameter: 310.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  rhinovirus A2 rhinovirus A2 |
| Molecular weight | Theoretical: 32.2741 KDa |
| Sequence | String: NPVENYIDEV LNEVLVVPNI NSSNPTTSNS APALDAAETG HTSSVQPEDV IETRYVQTSQ TRDEMSLESF LGRSGCIHES KLEVTLANY NKENFTVWAI NLQEMAQIRR KFELFTYTRF DSEITLVPCI SALSQDIGHI TMQYMYVPPG APVPNSRDDY A WQSGTNAS ...String: NPVENYIDEV LNEVLVVPNI NSSNPTTSNS APALDAAETG HTSSVQPEDV IETRYVQTSQ TRDEMSLESF LGRSGCIHES KLEVTLANY NKENFTVWAI NLQEMAQIRR KFELFTYTRF DSEITLVPCI SALSQDIGHI TMQYMYVPPG APVPNSRDDY A WQSGTNAS VFWQHGQAYP RFSLPFLSVA SAYYMFYDGY DEQDQNYGTA NTNNMGSLCS RIVTEKHIHK VHIMTRIYHK AK HVKAWCP RPPRALEYTR AHRTNFKIED RSIQTAIVTR PIITTA UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  rhinovirus A2 rhinovirus A2 |
| Molecular weight | Theoretical: 29.009588 KDa |
| Sequence | String: SPTVEACGYS DRIIQITRGD STITSQDVAN AIVAYGVWPH YLSSKDASAI DKPSQPDTSS NRFYTLRSVT WSSSSKGWWW KLPDALKDM GIFGENMFYH YLGRSGYTIH VQCNASKFHQ GTLIVALIPE HQIASALHGN VNVGYNYTHP GETGREVKAE T RLNPDLQP ...String: SPTVEACGYS DRIIQITRGD STITSQDVAN AIVAYGVWPH YLSSKDASAI DKPSQPDTSS NRFYTLRSVT WSSSSKGWWW KLPDALKDM GIFGENMFYH YLGRSGYTIH VQCNASKFHQ GTLIVALIPE HQIASALHGN VNVGYNYTHP GETGREVKAE T RLNPDLQP TEEYWLNFDG TLLGNITIFP HQFINLRSNN SATIIAPYVN AVPMDSMRSH NNWSLVIIPI CPLETSSAIN TI PITISIS PMCAEFSGAR AKRQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  rhinovirus A2 rhinovirus A2 |
| Molecular weight | Theoretical: 26.107793 KDa |
| Sequence | String: GLPVFITPGS GQFLTTDDFQ SPCALPWYHP TKEISIPGEV KNLVEICQVD SLVPINNTDT YINSENMYSV VLQSSINAPD KIFSIRTDV ASQPLATTLI GEISSYFTHW TGSLRFSFMF CGTANTTVKL LLAYTPPGIA EPTTRKDAML GTHVIWDVGL Q STISMVVP ...String: GLPVFITPGS GQFLTTDDFQ SPCALPWYHP TKEISIPGEV KNLVEICQVD SLVPINNTDT YINSENMYSV VLQSSINAPD KIFSIRTDV ASQPLATTLI GEISSYFTHW TGSLRFSFMF CGTANTTVKL LLAYTPPGIA EPTTRKDAML GTHVIWDVGL Q STISMVVP WISASHYRNT SPGRSTSGYI TCWYQTRLVI PPQTPPTARL LCFVSGCKDF CLRMARDTNL HLQSGAIAQ UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  rhinovirus A2 rhinovirus A2 |
| Molecular weight | Theoretical: 7.356971 KDa |
| Sequence | String: GAQVSRQNVG THSTQNSVSN GSSLNYFNIN YFKDAASNGA SKLEFTQDPS KFTDPVKDVL EKGIPTLQ UniProtKB: Genome polyprotein |
-Macromolecule #5: LAURIC ACID
| Macromolecule | Name: LAURIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: DAO |
|---|---|
| Molecular weight | Theoretical: 200.318 Da |
| Chemical component information |  ChemComp-DAO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.8 Component:
Details: 0.1 M phosphate buffer, pH 5.8 | |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 293 K / Instrument: LEICA EM GP | |||||||||
| Details | The virus sample was purified from HeLa cells, diluted to a final concentration of 1 mg/mL in 100 mM potassium phosphate buffer, pH 5.8, and incubated at four degrees for one hour before the vitrification. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 2 / Number real images: 246 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 100000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)