[English] 日本語
 Yorodumi
Yorodumi- EMDB-4896: Single particle cryo-EM reconstruction of a 20-mer assembly of re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4896 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo-EM reconstruction of a 20-mer assembly of reduced recombinant human alphaA-crystallin. | |||||||||
 Map data Map data | Single particle cryo-EM reconstruction of a 20-mer assembly of reduced recombinant human alphaA-crystallin. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
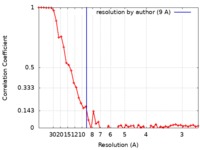
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Peters C / Kaiser CJO / Buchner J / Weinkauf S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: The structure and oxidation of the eye lens chaperone αA-crystallin. Authors: Christoph J O Kaiser / Carsten Peters / Philipp W N Schmid / Maria Stavropoulou / Juan Zou / Vinay Dahiya / Evgeny V Mymrikov / Beate Rockel / Sam Asami / Martin Haslbeck / Juri Rappsilber / ...Authors: Christoph J O Kaiser / Carsten Peters / Philipp W N Schmid / Maria Stavropoulou / Juan Zou / Vinay Dahiya / Evgeny V Mymrikov / Beate Rockel / Sam Asami / Martin Haslbeck / Juri Rappsilber / Bernd Reif / Martin Zacharias / Johannes Buchner / Sevil Weinkauf /   Abstract: The small heat shock protein αA-crystallin is a molecular chaperone important for the optical properties of the vertebrate eye lens. It forms heterogeneous oligomeric ensembles. We determined the ...The small heat shock protein αA-crystallin is a molecular chaperone important for the optical properties of the vertebrate eye lens. It forms heterogeneous oligomeric ensembles. We determined the structures of human αA-crystallin oligomers by combining cryo-electron microscopy, cross-linking/mass spectrometry, NMR spectroscopy and molecular modeling. The different oligomers can be interconverted by the addition or subtraction of tetramers, leading to mainly 12-, 16- and 20-meric assemblies in which interactions between N-terminal regions are important. Cross-dimer domain-swapping of the C-terminal region is a determinant of αA-crystallin heterogeneity. Human αA-crystallin contains two cysteines, which can form an intramolecular disulfide in vivo. Oxidation in vitro requires conformational changes and oligomer dissociation. The oxidized oligomers, which are larger than reduced αA-crystallin and destabilized against unfolding, are active chaperones and can transfer the disulfide to destabilized substrate proteins. The insight into the structure and function of αA-crystallin provides a basis for understanding its role in the eye lens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4896.map.gz emd_4896.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4896-v30.xml emd-4896-v30.xml emd-4896.xml emd-4896.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4896_fsc.xml emd_4896_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4896.png emd_4896.png | 117.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4896 http://ftp.pdbj.org/pub/emdb/structures/EMD-4896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4896 | HTTPS FTP |
-Validation report
| Summary document |  emd_4896_validation.pdf.gz emd_4896_validation.pdf.gz | 229.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4896_full_validation.pdf.gz emd_4896_full_validation.pdf.gz | 229.1 KB | Display | |
| Data in XML |  emd_4896_validation.xml.gz emd_4896_validation.xml.gz | 8.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4896 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4896 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4896 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4896 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4896.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4896.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
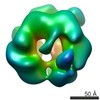
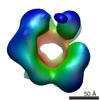
| Annotation | Single particle cryo-EM reconstruction of a 20-mer assembly of reduced recombinant human alphaA-crystallin. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Reduced recombinant human alphaA-crystallin
| Entire | Name: Reduced recombinant human alphaA-crystallin |
|---|---|
| Components |
|
-Supramolecule #1: Reduced recombinant human alphaA-crystallin
| Supramolecule | Name: Reduced recombinant human alphaA-crystallin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinant wild type full-length human alphaA-crystallin purified in the presence of reductant. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: eye / Tissue: lens / Location in cell: cytoplasm Homo sapiens (human) / Organ: eye / Tissue: lens / Location in cell: cytoplasm |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 398 KDa |
-Macromolecule #1: alphaA-crystallin
| Macromolecule | Name: alphaA-crystallin / type: protein_or_peptide / ID: 1 / Details: Wild type full sequence / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: eye / Tissue: lens Homo sapiens (human) / Organ: eye / Tissue: lens |
| Recombinant expression | Organism:  |
| Sequence | String: MDVTIQHPWF KRTLGPFYPS RLFDQFFGEG LFEYDLLPFL SSTISPYYRQ SLFRTVLDSG ISEVRSDRD KFVIFLDVKH FSPEDLTVKV QDDFVEIHGK HNERQDDHGY ISREFHRRYR L PSNVDQSA LSCSLSADGM LTFCGPKIQT GLDATHAERA IPVSREEKPT SAPSS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Buffer was prepared without EDTA and DTT. EDTA stock (500mM) was titrated to pH 8 and added to 1 mM. DTT stock (1M) was added to 1mM. | |||||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.007 kPa | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 293 K / Instrument: HOMEMADE PLUNGER Details: Diluted equilibrated specimen was added to glow-discharged (30s) grids. Sample was blotted 30s after sample application and immediately plunged.. | |||||||||||||||||||||
| Details | Specimen was thawed, diluted to the final concentration and equilibrated at 310K for 3h. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-10 / Number grids imaged: 3 / Average exposure time: 3.2 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 37000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)