[English] 日本語
 Yorodumi
Yorodumi- EMDB-6218: Yeast Rvb1 and Rvb2 proteins oligomerize as a conformationally va... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6218 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
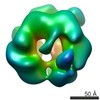
| Title | Yeast Rvb1 and Rvb2 proteins oligomerize as a conformationally variable dodecamer with low frequency | |||||||||
 Map data Map data | In vivo assembled yeast Rvb1/Rvb2 complex (dodecamer, CL4) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ / Electron microscopy / Pontin / Reptin / Chromatin remodeling | |||||||||
| Function / homology |  Function and homology information Function and homology informationTTT Hsp90 cochaperone complex / R2TP complex / Swr1 complex / Ino80 complex / box C/D snoRNP assembly / 3'-5' DNA helicase activity / NuA4 histone acetyltransferase complex / DNA helicase activity / rRNA processing / 5'-3' DNA helicase activity ...TTT Hsp90 cochaperone complex / R2TP complex / Swr1 complex / Ino80 complex / box C/D snoRNP assembly / 3'-5' DNA helicase activity / NuA4 histone acetyltransferase complex / DNA helicase activity / rRNA processing / 5'-3' DNA helicase activity / DNA helicase / protein stabilization / chromatin remodeling / DNA repair / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / chromatin / ATP hydrolysis activity / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Jeganathan A / Leong A / Huen J / Zhao L / Nano N / Houry WA / Ortega J | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2015 Journal: J Mol Biol / Year: 2015Title: Yeast rvb1 and rvb2 proteins oligomerize as a conformationally variable dodecamer with low frequency. Authors: Ajitha Jeganathan / Vivian Leong / Liang Zhao / Jennifer Huen / Nardin Nano / Walid A Houry / Joaquin Ortega /  Abstract: Rvb1 and Rvb2 are conserved AAA+ (ATPases associated with diverse cellular activities) proteins found at the core of large multicomponent complexes that play key roles in chromatin remodeling, ...Rvb1 and Rvb2 are conserved AAA+ (ATPases associated with diverse cellular activities) proteins found at the core of large multicomponent complexes that play key roles in chromatin remodeling, integrity of the telomeres, ribonucleoprotein complex biogenesis and other essential cellular processes. These proteins contain an AAA+ domain for ATP binding and hydrolysis and an insertion domain proposed to bind DNA/RNA. Yeast Rvb1 and Rvb2 proteins oligomerize primarily as heterohexameric rings. The six AAA+ core domains form the body of the ring and the insertion domains protrude from one face of the ring. Conversely, human Rvbs form a mixture of hexamers and dodecamers made of two stacked hexamers interacting through the insertion domains. Human dodecamers adopt either a contracted or a stretched conformation. Here, we found that yeast Rvb1/Rvb2 complexes when assembled in vivo mainly form hexamers but they also assemble as dodecamers with a frequency lower than 10%. Yeast dodecamers adopt not only the stretched and contracted structures that have been described for human Rvb1/Rvb2 dodecamers but also intermediate conformations in between these two extreme states. The orientation of the insertion domains of Rvb1 and Rvb2 proteins in these conformers changes as the dodecamer transitions from the stretched structure to a more contracted structure. Finally, we observed that for the yeast proteins, oligomerization as a dodecamer inhibits the ATPase activity of the Rvb1/Rvb2 complex. These results indicate that although human and yeast Rvb1 and Rvb2 proteins share high degree of homology, there are significant differences in their oligomeric behavior and dynamics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6218.map.gz emd_6218.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6218-v30.xml emd-6218-v30.xml emd-6218.xml emd-6218.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6218.png emd_6218.png | 217.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6218 http://ftp.pdbj.org/pub/emdb/structures/EMD-6218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6218 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6218.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6218.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | In vivo assembled yeast Rvb1/Rvb2 complex (dodecamer, CL4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.54 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : In vivo assembled Rvb1/Rvb2 complex, CL4
| Entire | Name: In vivo assembled Rvb1/Rvb2 complex, CL4 |
|---|---|
| Components |
|
-Supramolecule #1000: In vivo assembled Rvb1/Rvb2 complex, CL4
| Supramolecule | Name: In vivo assembled Rvb1/Rvb2 complex, CL4 / type: sample / ID: 1000 Oligomeric state: 6 molecules of Rvb1 and 6 molecules of Rvb2 Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 600 KDa / Theoretical: 600 KDa / Method: Size exclusion chromatography |
-Macromolecule #1: Rvb1
| Macromolecule | Name: Rvb1 / type: protein_or_peptide / ID: 1 Name.synonym: RuvBL1, Tip49a, Tip48 / Tip49, ECP-54, Pontin, Tih1, p50, Tap54b Number of copies: 6 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 50.5 KDa / Theoretical: 50.5 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: RuvB-like protein 1 |
-Macromolecule #2: Rvb2
| Macromolecule | Name: Rvb2 / type: protein_or_peptide / ID: 2 Name.synonym: RuvBL2, Tip49b, Tip49 / Tip48, ECP-51, Reptin, Tih2, p47, Tap54a Number of copies: 6 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 51.6 KDa / Theoretical: 51.6 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: RuvB-like protein 2 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 25 mM Tris-HCl, pH 7.5, 80 mM KCl, 10% v/v glycerol, 1 mM DTT |
|---|---|
| Staining | Type: NEGATIVE / Details: 2% uranyl acetate |
| Grid | Details: 400 mesh grids with thin carbon support, glow-discharged in air atmosphere |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Date | Feb 1, 2014 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 12.7 µm / Number real images: 100 / Average electron dose: 15 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: JEOL |
- Image processing
Image processing
| Details | Particles selected with boxer, image classification done using ML3D, and projection matching 3D reconstruction done with Xmipp. |
|---|---|
| CTF correction | Details: CTFFIND3 |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: Xmipp / Number images used: 1718 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















