[English] 日本語
 Yorodumi
Yorodumi- EMDB-48906: Azotobacter vinelandii extended type VI secretion system sheath t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Azotobacter vinelandii extended type VI secretion system sheath tube complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | secretion system / TRANSPORT PROTEIN | |||||||||
| Function / homology | Type VI secretion system TssC-like / TssC1, N-terminal / TssC1, C-terminal / EvpB/VC_A0108, tail sheath N-terminal domain / EvpB/VC_A0108, tail sheath gpW/gp25-like domain / Type VI secretion system sheath protein TssB1 / Type VI secretion system, VipA, VC_A0107 or Hcp2 / Type VI secretion system contractile sheath small subunit / DUF877 family protein Function and homology information Function and homology information | |||||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 1.85 Å | |||||||||
 Authors Authors | Warmack RA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2025 Journal: Structure / Year: 2025Title: CryoEM-enabled visual proteomics reveals de novo structures of oligomeric protein complexes. Authors: Yuanbo Shen / Ailiena O Maggiolo / Tianzheng Zhang / Rebeccah A Warmack /  Abstract: Single particle cryoelectron microscopy (cryoEM) and cryoelectron tomography (cryoET) are powerful methods for unveiling unique and functionally relevant structural states. Aided by mass spectrometry ...Single particle cryoelectron microscopy (cryoEM) and cryoelectron tomography (cryoET) are powerful methods for unveiling unique and functionally relevant structural states. Aided by mass spectrometry and machine learning, they promise to facilitate the visual exploration of proteomes. Leveraging visual proteomics, we interrogate structures isolated from a complex cellular milieu by cryoEM to identify and classify molecular structures and complexes de novo. By comparing three automated model building programs, CryoID, DeepTracer, and ModelAngelo, we determine the identity of six distinct oligomeric protein complexes from partially purified extracts of the nitrogen-fixing bacterium Azotobacter vinelandii using both anaerobic and aerobic cryoEM, including two original oligomeric structures. Overall, by allowing the study of near-native oligomeric protein states, cryoEM-enabled visual proteomics reveals unique structures that correspond to relevant species observed in situ. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48906.map.gz emd_48906.map.gz | 2.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48906-v30.xml emd-48906-v30.xml emd-48906.xml emd-48906.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_48906_fsc.xml emd_48906_fsc.xml | 28.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_48906.png emd_48906.png | 117.8 KB | ||
| Filedesc metadata |  emd-48906.cif.gz emd-48906.cif.gz | 5.8 KB | ||
| Others |  emd_48906_half_map_1.map.gz emd_48906_half_map_1.map.gz emd_48906_half_map_2.map.gz emd_48906_half_map_2.map.gz | 2.2 GB 2.2 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48906 http://ftp.pdbj.org/pub/emdb/structures/EMD-48906 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48906 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48906 | HTTPS FTP |
-Related structure data
| Related structure data |  9n4vMC  9n4wC  9n4xC  9n4yC  9n59C  9n5aC  9nsvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_48906.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48906.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_48906_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
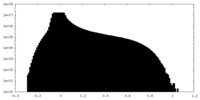
| Projections & Slices |
| ||||||||||||
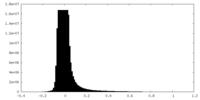
| Density Histograms |
-Half map: #2
| File | emd_48906_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
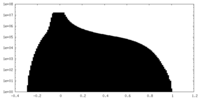
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Filamentous assembly of the extended type VI secretion system she...
| Entire | Name: Filamentous assembly of the extended type VI secretion system sheath tube complex |
|---|---|
| Components |
|
-Supramolecule #1: Filamentous assembly of the extended type VI secretion system she...
| Supramolecule | Name: Filamentous assembly of the extended type VI secretion system sheath tube complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) / Strain: DJ Azotobacter vinelandii (bacteria) / Strain: DJ |
-Macromolecule #1: DUF877 family protein
| Macromolecule | Name: DUF877 family protein / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) / Strain: DJ Azotobacter vinelandii (bacteria) / Strain: DJ |
| Molecular weight | Theoretical: 55.032344 KDa |
| Sequence | String: MAQPSAGETQ ASENITLSLL DRIIAEGRMA HDDSQQDYAR DMLAEFATQV LDEGMAIDKD TVSMINDRIA QIDALIGAQL DEILHHPEL QKLEASWRGL HMLVKNTETG ARLKLRLLNV TQKELLIDLE KAVEFDQSAL FKKIYEEEYG TFGGHPFSLL V GDYSFGRH ...String: MAQPSAGETQ ASENITLSLL DRIIAEGRMA HDDSQQDYAR DMLAEFATQV LDEGMAIDKD TVSMINDRIA QIDALIGAQL DEILHHPEL QKLEASWRGL HMLVKNTETG ARLKLRLLNV TQKELLIDLE KAVEFDQSAL FKKIYEEEYG TFGGHPFSLL V GDYSFGRH PQDIGLLEKL SNVAAAAHAP FIAAASPRLF DMGSFTELAV PRDLAKIFES QELIKWRAFR ESEDSRYVSL VL PHVLLRL PYGPDTCPVE GMDYVEDVNG RDHARYLWGN AAWALTQRIT EAFARYGWCA AIRGVEGGGA VEGLPAHSFR TSS GDLSLK CPTEVAITDR REKELDALGF IALCHKKNSD LAVFFGSQTT NRPRVYNTNE ANANARISAM LPYVLAASRF AHYL KVIMR DKVGSFMTRD NVQTYLNNWI ADYVLINDNA PQEIKAQYPL REARVDVSEV VGKPGVYRAT VFLRPHFQLE ELTAS IRLV ATLPPPAAA UniProtKB: DUF877 family protein |
-Macromolecule #2: Type VI secretion system contractile sheath small subunit
| Macromolecule | Name: Type VI secretion system contractile sheath small subunit type: protein_or_peptide / ID: 2 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) / Strain: DJ Azotobacter vinelandii (bacteria) / Strain: DJ |
| Molecular weight | Theoretical: 20.849465 KDa |
| Sequence | String: MAESTQHKLD RIRPPRVQIT YDVETGNAIE KKELPLVVGI LADLSGKPEK PLPKLMERRF VEINRDNFND VLASIAPRAA LQVDNTLSQ DGSKLNIELH FDHIDDFDPV NIVRQVTPLR RLFEARQRLR DLLTKLDGND DLDKLLQDVV ANTEGLQEIR S ARPQAENP AGAPGAEPAA DEPAAEPQA UniProtKB: Type VI secretion system contractile sheath small subunit |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 6150 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -2.5 µm / Nominal defocus min: -0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)