[English] 日本語
 Yorodumi
Yorodumi- EMDB-4791: full-length bacterial polysaccharide co-polymerase - C1 symmetry -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4791 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | full-length bacterial polysaccharide co-polymerase - C1 symmetry | |||||||||
 Map data Map data | Full-length bacterial polysaccharide co-polymerase - C1 symmetry | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
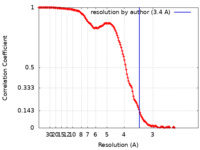
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Wiseman B / Nitharwal RG / Hogbom M | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structure of a full-length bacterial polysaccharide co-polymerase. Authors: Benjamin Wiseman / Ram Gopal Nitharwal / Göran Widmalm / Martin Högbom /   Abstract: Lipopolysaccharides are important components of the bacterial cell envelope that among other things act as a protective barrier against the environment and toxic molecules such as antibiotics. One of ...Lipopolysaccharides are important components of the bacterial cell envelope that among other things act as a protective barrier against the environment and toxic molecules such as antibiotics. One of the most widely disseminated pathways of polysaccharide biosynthesis is the inner membrane bound Wzy-dependent pathway. Here we present the 3.0 Å structure of the co-polymerase component of this pathway, WzzB from E. coli solved by single-particle cryo-electron microscopy. The overall architecture is octameric and resembles a box jellyfish containing a large bell-shaped periplasmic domain with the 2-helix transmembrane domain from each protomer, positioned 32 Å apart, encircling a large empty transmembrane chamber. This structure also reveals the architecture of the transmembrane domain, including the location of key residues for the Wzz-family of proteins and the Wzy-dependent pathway present in many Gram-negative bacteria, explaining several of the previous biochemical and mutational studies and lays the foundation for future investigations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4791.map.gz emd_4791.map.gz | 8.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4791-v30.xml emd-4791-v30.xml emd-4791.xml emd-4791.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4791_fsc.xml emd_4791_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4791.png emd_4791.png | 58.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4791 http://ftp.pdbj.org/pub/emdb/structures/EMD-4791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4791 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4791.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4791.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full-length bacterial polysaccharide co-polymerase - C1 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Octameric bacterial polysaccharide co-polymerase complex
| Entire | Name: Octameric bacterial polysaccharide co-polymerase complex |
|---|---|
| Components |
|
-Supramolecule #1: Octameric bacterial polysaccharide co-polymerase complex
| Supramolecule | Name: Octameric bacterial polysaccharide co-polymerase complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Bacterial polysaccharide co-polymerase
| Macromolecule | Name: Bacterial polysaccharide co-polymerase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MRVENNNVSG QNHDPEQIDL IDLLVQLWRG KMTIIISVIV AIALAIGYLA VAKEKWTSTA IITQPDVGQI AGYNNAMNVI YGQAAPKVSD LQETLIGRFS SAFSALAETL DNQEEREKLT IEPSVKNQQL PLTVSYVGQT AEGAQMKLAQ YIQQVDDKVN QELEKDLKDN ...String: MRVENNNVSG QNHDPEQIDL IDLLVQLWRG KMTIIISVIV AIALAIGYLA VAKEKWTSTA IITQPDVGQI AGYNNAMNVI YGQAAPKVSD LQETLIGRFS SAFSALAETL DNQEEREKLT IEPSVKNQQL PLTVSYVGQT AEGAQMKLAQ YIQQVDDKVN QELEKDLKDN IALGRKNLQD SLRTQEVVAQ EQKDLRIRQI QEALQYANQA QVTKPQIQQT GEDITQDTLF LLGSEALESM IKHEATRPLV FSPNYYQTRQ NLLDIESLKV DDLDIHAYRY VMKPMLPIRR DSPKKAITLI LAVLLGGMVG AGIVLGRNAL RNYNAKEFRV PGSHHHHHHH H |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Details: 40 seconds at 20 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2347 / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)