[English] 日本語
 Yorodumi
Yorodumi- EMDB-4798: full-length bacterial polysaccharide co-polymerase - C8 symmetry -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4798 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | full-length bacterial polysaccharide co-polymerase - C8 symmetry | |||||||||
 Map data Map data | full-length bacterial polysaccharide co-polymerase - C8 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / enzyme / polysaccharide / co-polymerase | |||||||||
| Function / homology | Polysaccharide chain length determinant N-terminal domain / Chain length determinant protein / : / lipopolysaccharide biosynthetic process / protein tyrosine kinase activity / plasma membrane / Chain length determinant protein Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
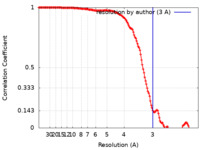
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Wiseman B / Nitharwal RG | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structure of a full-length bacterial polysaccharide co-polymerase. Authors: Benjamin Wiseman / Ram Gopal Nitharwal / Göran Widmalm / Martin Högbom /   Abstract: Lipopolysaccharides are important components of the bacterial cell envelope that among other things act as a protective barrier against the environment and toxic molecules such as antibiotics. One of ...Lipopolysaccharides are important components of the bacterial cell envelope that among other things act as a protective barrier against the environment and toxic molecules such as antibiotics. One of the most widely disseminated pathways of polysaccharide biosynthesis is the inner membrane bound Wzy-dependent pathway. Here we present the 3.0 Å structure of the co-polymerase component of this pathway, WzzB from E. coli solved by single-particle cryo-electron microscopy. The overall architecture is octameric and resembles a box jellyfish containing a large bell-shaped periplasmic domain with the 2-helix transmembrane domain from each protomer, positioned 32 Å apart, encircling a large empty transmembrane chamber. This structure also reveals the architecture of the transmembrane domain, including the location of key residues for the Wzz-family of proteins and the Wzy-dependent pathway present in many Gram-negative bacteria, explaining several of the previous biochemical and mutational studies and lays the foundation for future investigations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4798.map.gz emd_4798.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4798-v30.xml emd-4798-v30.xml emd-4798.xml emd-4798.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4798_fsc.xml emd_4798_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4798.png emd_4798.png | 64.1 KB | ||
| Filedesc metadata |  emd-4798.cif.gz emd-4798.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4798 http://ftp.pdbj.org/pub/emdb/structures/EMD-4798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4798 | HTTPS FTP |
-Related structure data
| Related structure data |  6rbgMC  4791C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4798.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4798.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full-length bacterial polysaccharide co-polymerase - C8 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Octameric bacterial polysaccharide co-polymerase complex
| Entire | Name: Octameric bacterial polysaccharide co-polymerase complex |
|---|---|
| Components |
|
-Supramolecule #1: Octameric bacterial polysaccharide co-polymerase complex
| Supramolecule | Name: Octameric bacterial polysaccharide co-polymerase complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Chain length determinant protein
| Macromolecule | Name: Chain length determinant protein / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.370621 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRVENNNVSG QNHDPEQIDL IDLLVQLWRG KMTIIISVIV AIALAIGYLA VAKEKWTSTA IITQPDVGQI AGYNNAMNVI YGQAAPKVS DLQETLIGRF SSAFSALAET LDNQEEREKL TIEPSVKNQQ LPLTVSYVGQ TAEGAQMKLA QYIQQVDDKV N QELEKDLK ...String: MRVENNNVSG QNHDPEQIDL IDLLVQLWRG KMTIIISVIV AIALAIGYLA VAKEKWTSTA IITQPDVGQI AGYNNAMNVI YGQAAPKVS DLQETLIGRF SSAFSALAET LDNQEEREKL TIEPSVKNQQ LPLTVSYVGQ TAEGAQMKLA QYIQQVDDKV N QELEKDLK DNIALGRKNL QDSLRTQEVV AQEQKDLRIR QIQEALQYAN QAQVTKPQIQ QTGEDITQDT LFLLGSEALE SM IKHEATR PLVFSPNYYQ TRQNLLDIES LKVDDLDIHA YRYVMKPMLP IRRDSPKKAI TLILAVLLGG MVGAGIVLGR NAL RNYNAK EFRVPGSHHH HHHHH UniProtKB: Chain length determinant protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Details: 40 seconds at 20 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2347 / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 87 |
|---|---|
| Output model |  PDB-6rbg: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)