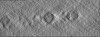[English] 日本語
 Yorodumi
Yorodumi- EMDB-44847: Tomograms of isolated synaptic vesicles from Syp-/- mouse brain -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tomograms of isolated synaptic vesicles from Syp-/- mouse brain | |||||||||
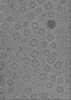
 Map data Map data | tomogram of Syp-/- isolated synaptic vesicles | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | V-ATPase / synaptic vesicle / MEMBRANE PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Wang C / Jiang W / Yang K / Wang X / Guo Q / Brunger AT | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structure and topography of the synaptic V-ATPase-synaptophysin complex. Authors: Chuchu Wang / Wenhong Jiang / Jeremy Leitz / Kailu Yang / Luis Esquivies / Xing Wang / Xiaotao Shen / Richard G Held / Daniel J Adams / Tamara Basta / Lucas Hampton / Ruiqi Jian / Lihua ...Authors: Chuchu Wang / Wenhong Jiang / Jeremy Leitz / Kailu Yang / Luis Esquivies / Xing Wang / Xiaotao Shen / Richard G Held / Daniel J Adams / Tamara Basta / Lucas Hampton / Ruiqi Jian / Lihua Jiang / Michael H B Stowell / Wolfgang Baumeister / Qiang Guo / Axel T Brunger /     Abstract: Synaptic vesicles are organelles with a precisely defined protein and lipid composition, yet the molecular mechanisms for the biogenesis of synaptic vesicles are mainly unknown. Here we discovered a ...Synaptic vesicles are organelles with a precisely defined protein and lipid composition, yet the molecular mechanisms for the biogenesis of synaptic vesicles are mainly unknown. Here we discovered a well-defined interface between the synaptic vesicle V-ATPase and synaptophysin by in situ cryo-electron tomography and single-particle cryo-electron microscopy of functional synaptic vesicles isolated from mouse brains. The synaptic vesicle V-ATPase is an ATP-dependent proton pump that establishes the proton gradient across the synaptic vesicle, which in turn drives the uptake of neurotransmitters. Synaptophysin and its paralogues synaptoporin and synaptogyrin belong to a family of abundant synaptic vesicle proteins whose function is still unclear. We performed structural and functional studies of synaptophysin-knockout mice, confirming the identity of synaptophysin as an interaction partner with the V-ATPase. Although there is little change in the conformation of the V-ATPase upon interaction with synaptophysin, the presence of synaptophysin in synaptic vesicles profoundly affects the copy number of V-ATPases. This effect on the topography of synaptic vesicles suggests that synaptophysin assists in their biogenesis. In support of this model, we observed that synaptophysin-knockout mice exhibit severe seizure susceptibility, suggesting an imbalance of neurotransmitter release as a physiological consequence of the absence of synaptophysin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44847.map.gz emd_44847.map.gz | 242.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44847-v30.xml emd-44847-v30.xml emd-44847.xml emd-44847.xml | 30.8 KB 30.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44847.png emd_44847.png | 165.2 KB | ||
| Filedesc metadata |  emd-44847.cif.gz emd-44847.cif.gz | 4.4 KB | ||
| Others |  emd_44847_additional_1.map.gz emd_44847_additional_1.map.gz emd_44847_additional_10.map.gz emd_44847_additional_10.map.gz emd_44847_additional_2.map.gz emd_44847_additional_2.map.gz emd_44847_additional_3.map.gz emd_44847_additional_3.map.gz emd_44847_additional_4.map.gz emd_44847_additional_4.map.gz emd_44847_additional_5.map.gz emd_44847_additional_5.map.gz emd_44847_additional_6.map.gz emd_44847_additional_6.map.gz emd_44847_additional_7.map.gz emd_44847_additional_7.map.gz emd_44847_additional_8.map.gz emd_44847_additional_8.map.gz emd_44847_additional_9.map.gz emd_44847_additional_9.map.gz | 242.3 MB 242.2 MB 242.2 MB 242.2 MB 242.3 MB 242.4 MB 242.3 MB 242.2 MB 242.2 MB 242.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44847 http://ftp.pdbj.org/pub/emdb/structures/EMD-44847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44847 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44847.map.gz / Format: CCP4 / Size: 262.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44847.map.gz / Format: CCP4 / Size: 262.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | tomogram of Syp-/- isolated synaptic vesicles | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.888 Å | ||||||||||||||||||||||||||||||||
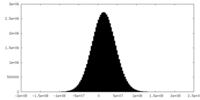
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
+Additional map: tomogram of Syp-/- isolated synaptic vesicles
- Sample components
Sample components
-Entire : Mouse brain isolated glutamatergic synaptic vesicles
| Entire | Name: Mouse brain isolated glutamatergic synaptic vesicles |
|---|---|
| Components |
|
-Supramolecule #1: Mouse brain isolated glutamatergic synaptic vesicles
| Supramolecule | Name: Mouse brain isolated glutamatergic synaptic vesicles / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#9 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | The specimen state should be an intact subcellular component. |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 41 |
|---|
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)